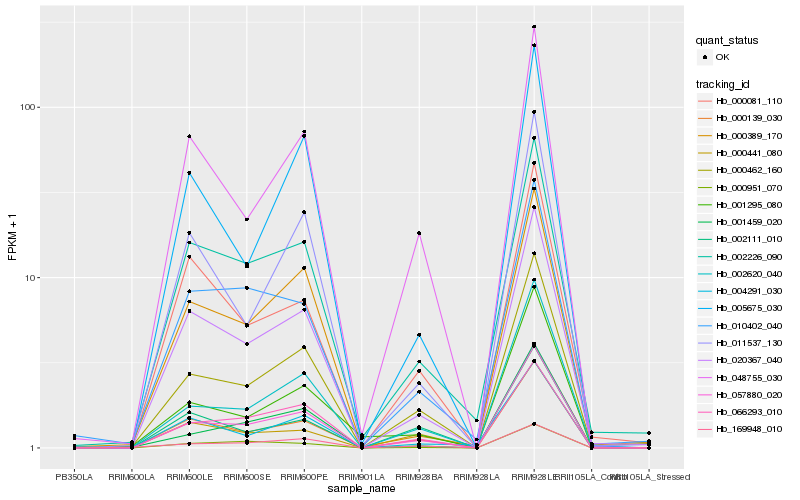
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_057880_020 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 2 |
Hb_000462_160 |
0.0661598164 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_020367_040 |
0.0764399029 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 4 |
Hb_002620_040 |
0.0773322017 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 5 |
Hb_000139_030 |
0.0838801679 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 6 |
Hb_048755_030 |
0.0991982836 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_066293_010 |
0.1014270361 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 8 |
Hb_002226_090 |
0.1036302256 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 9 |
Hb_001459_020 |
0.106691444 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 10 |
Hb_000951_070 |
0.1137938879 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 11 |
Hb_010402_040 |
0.1150332321 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 12 |
Hb_000389_170 |
0.1160464118 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 13 |
Hb_004291_030 |
0.1182185406 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 14 |
Hb_011537_130 |
0.1187344793 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_001295_080 |
0.1188605919 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 16 |
Hb_000441_080 |
0.1210215528 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_005675_030 |
0.121096343 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 18 |
Hb_169948_010 |
0.122669778 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 19 |
Hb_002111_010 |
0.1277971712 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 20 |
Hb_000081_110 |
0.1284189421 |
- |
- |
nitrate transporter, putative [Ricinus communis] |