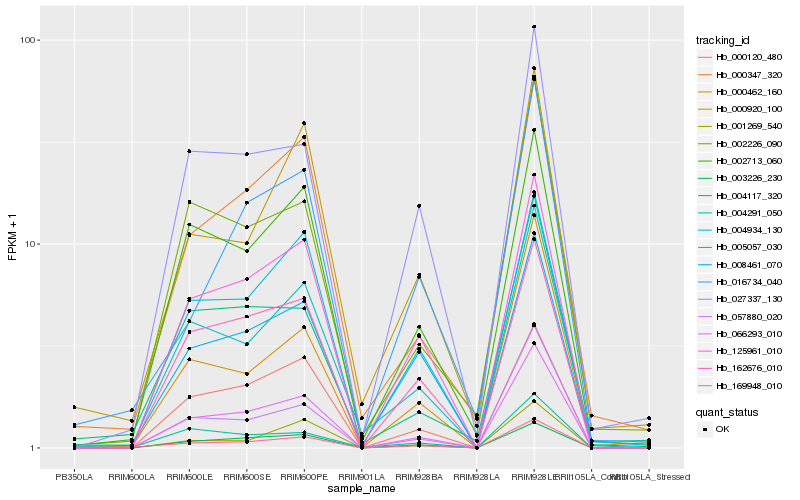
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169948_010 |
0.0 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 2 |
Hb_066293_010 |
0.0898545355 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 3 |
Hb_125961_010 |
0.1135633372 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_004934_130 |
0.1215652421 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 5 |
Hb_057880_020 |
0.122669778 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 6 |
Hb_000462_160 |
0.1245654102 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_027337_130 |
0.130602454 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 8 |
Hb_008461_070 |
0.1310176272 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003226_230 |
0.134071823 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 10 |
Hb_016734_040 |
0.1351462679 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 11 |
Hb_004117_320 |
0.1362639257 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 12 |
Hb_000347_320 |
0.136324495 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 13 |
Hb_002713_060 |
0.1367413758 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 14 |
Hb_004291_050 |
0.1380344945 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 15 |
Hb_000920_100 |
0.1400516257 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000120_480 |
0.1406618816 |
- |
- |
- |
| 17 |
Hb_001269_540 |
0.1415184237 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 18 |
Hb_162676_010 |
0.1419640836 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_002226_090 |
0.1439409639 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 20 |
Hb_005057_030 |
0.1443117604 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |