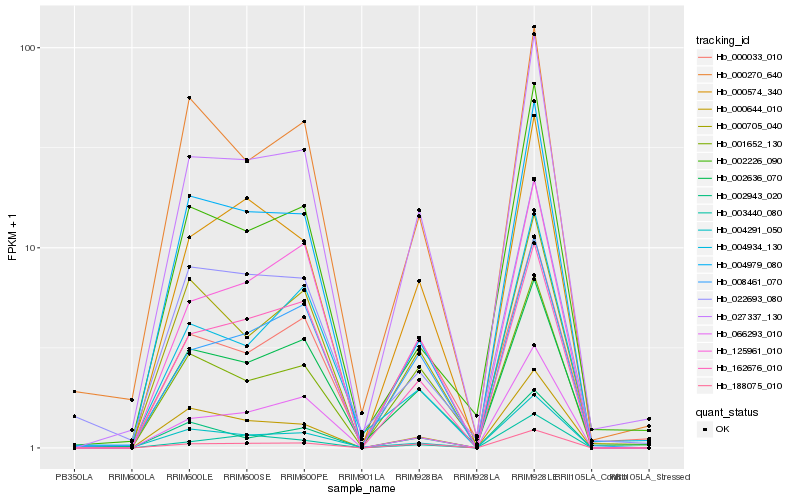
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027337_130 |
0.0 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 2 |
Hb_188075_010 |
0.0811176388 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 3 |
Hb_002636_070 |
0.0931337159 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 4 |
Hb_000574_340 |
0.0962599378 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 5 |
Hb_004291_050 |
0.0964220742 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 6 |
Hb_008461_070 |
0.0974033754 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000033_010 |
0.1057964346 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 8 |
Hb_125961_010 |
0.1079082583 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000644_010 |
0.1082483883 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001652_130 |
0.1096626654 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 11 |
Hb_004979_080 |
0.1111793076 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_000705_040 |
0.1115395311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_162676_010 |
0.1116858842 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_066293_010 |
0.1121546608 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 15 |
Hb_002226_090 |
0.1136140533 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 16 |
Hb_000270_640 |
0.1143734227 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 17 |
Hb_002943_020 |
0.1183166215 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 18 |
Hb_003440_080 |
0.1187140666 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 19 |
Hb_004934_130 |
0.1205872747 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 20 |
Hb_022693_080 |
0.1207182016 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |