| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
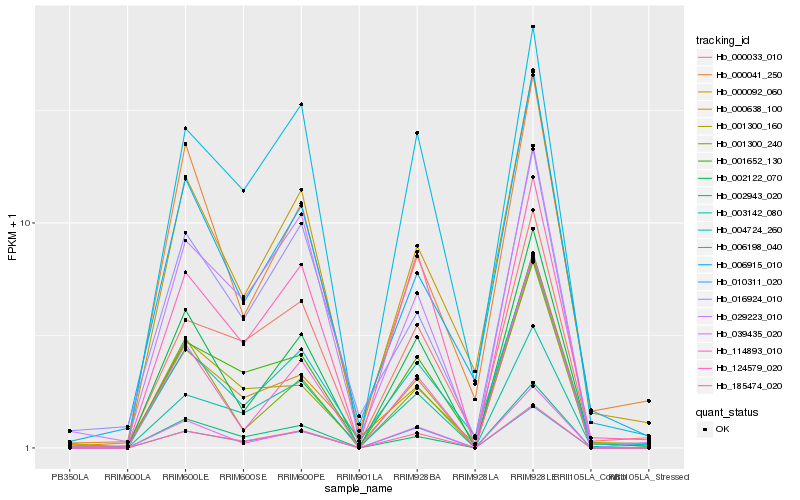
Hb_004724_260 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 2 |
Hb_000638_100 |
0.0706335637 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 3 |
Hb_000033_010 |
0.0987848889 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 4 |
Hb_006915_010 |
0.1072045755 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_006198_040 |
0.1092182979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002122_070 |
0.1118219244 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001652_130 |
0.113063147 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 8 |
Hb_185474_020 |
0.1187893708 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 9 |
Hb_003142_080 |
0.1238634068 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 10 |
Hb_124579_020 |
0.1247563785 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 11 |
Hb_000092_060 |
0.127420291 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 12 |
Hb_000041_250 |
0.1275954246 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 13 |
Hb_039435_020 |
0.1315970444 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_029223_010 |
0.1372833356 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 15 |
Hb_114893_010 |
0.1374735574 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 16 |
Hb_002943_020 |
0.1407060702 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 17 |
Hb_016924_010 |
0.1422576119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001300_240 |
0.1431948823 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_010311_020 |
0.1434576105 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_001300_160 |
0.1458664759 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |