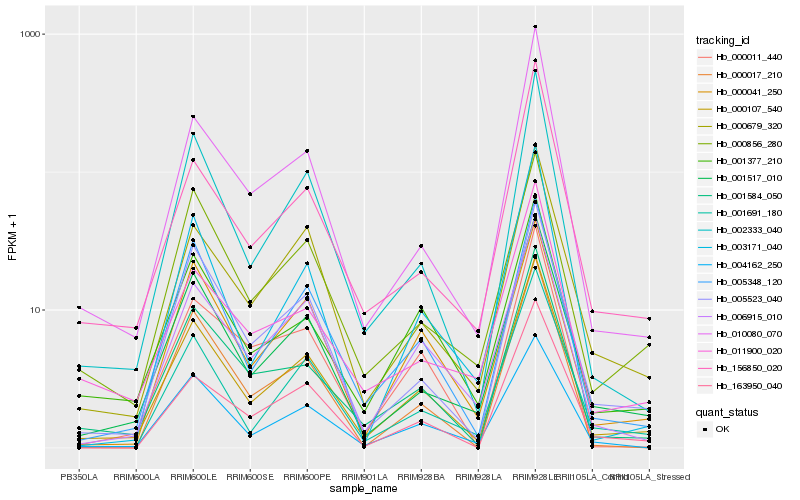
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_540 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 2 |
Hb_001517_010 |
0.0812117588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002333_040 |
0.0838726868 |
- |
- |
hypothetical protein CISIN_1g015895mg [Citrus sinensis] |
| 4 |
Hb_001584_050 |
0.0872622132 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005348_120 |
0.0904701329 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000679_320 |
0.0954862988 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 7 |
Hb_004162_250 |
0.0968207298 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 8 |
Hb_006915_010 |
0.0973880992 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_010080_070 |
0.0989021825 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001691_180 |
0.0989761858 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 11 |
Hb_001377_210 |
0.1016547046 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 12 |
Hb_005523_040 |
0.1040141274 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 13 |
Hb_156850_020 |
0.1085572203 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 14 |
Hb_163950_040 |
0.1116486104 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003171_040 |
0.1135062489 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 16 |
Hb_000041_250 |
0.1140616422 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 17 |
Hb_000856_280 |
0.1223327152 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 18 |
Hb_000011_440 |
0.1233262137 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 19 |
Hb_011900_020 |
0.123517526 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000017_210 |
0.1245606261 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |