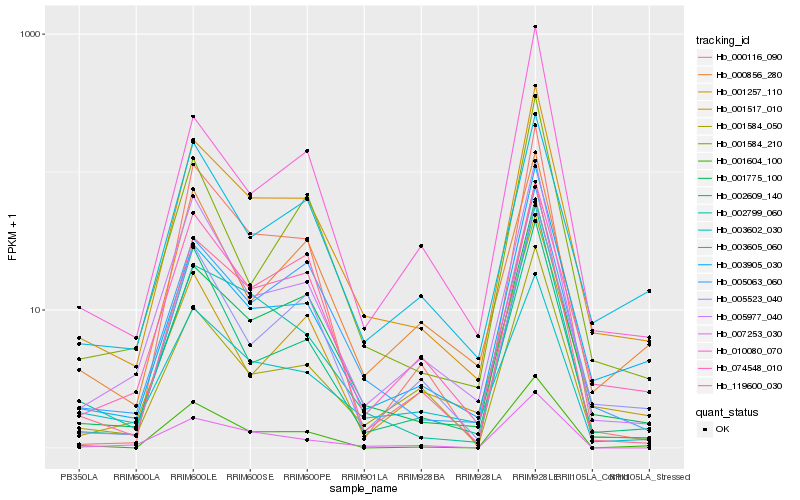
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001257_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636396 [Jatropha curcas] |
| 2 |
Hb_001775_100 |
0.0643521239 |
- |
- |
PREDICTED: probable 2-carboxy-D-arabinitol-1-phosphatase [Jatropha curcas] |
| 3 |
Hb_005063_060 |
0.0921854153 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 4 |
Hb_074548_010 |
0.0977778912 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_002799_060 |
0.0988696612 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_005523_040 |
0.1108832555 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 7 |
Hb_003905_030 |
0.1112096626 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 8 |
Hb_005977_040 |
0.1120314928 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 9 |
Hb_001584_050 |
0.1131554659 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001604_100 |
0.1254866135 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 11 |
Hb_003605_060 |
0.1259644953 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 12 |
Hb_119600_030 |
0.1272448578 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 13 |
Hb_003602_030 |
0.1275140296 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 14 |
Hb_001584_210 |
0.1300492781 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000116_090 |
0.1314354641 |
- |
- |
catalase [Hevea brasiliensis] |
| 16 |
Hb_007253_030 |
0.1326928879 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_000856_280 |
0.1335592441 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 18 |
Hb_001517_010 |
0.1335933684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010080_070 |
0.1356987674 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002609_140 |
0.1360993549 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |