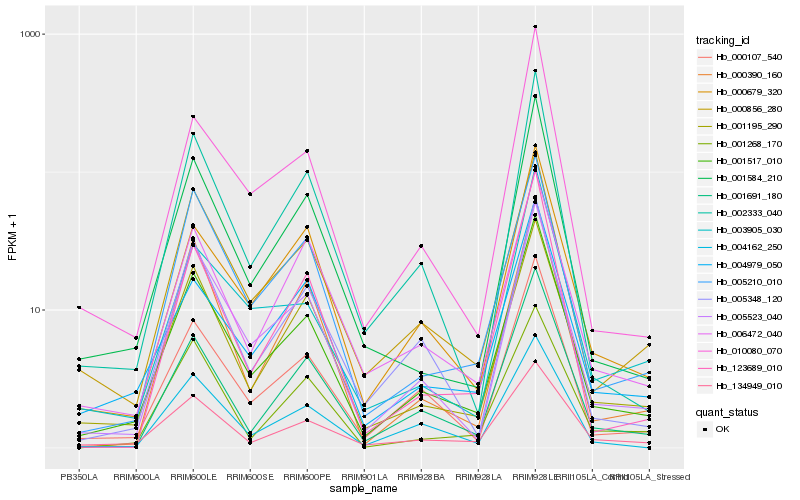
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001517_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001195_290 |
0.0797325797 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 3 |
Hb_000107_540 |
0.0812117588 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 4 |
Hb_005523_040 |
0.0824998431 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 5 |
Hb_134949_010 |
0.0928292473 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 6 |
Hb_001584_210 |
0.0935576276 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001691_180 |
0.095452275 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 8 |
Hb_000679_320 |
0.0980380738 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 9 |
Hb_002333_040 |
0.1015207538 |
- |
- |
hypothetical protein CISIN_1g015895mg [Citrus sinensis] |
| 10 |
Hb_004979_050 |
0.102835289 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 11 |
Hb_000856_280 |
0.1034903339 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 12 |
Hb_005210_010 |
0.1037118731 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 13 |
Hb_004162_250 |
0.1080568145 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 14 |
Hb_006472_040 |
0.1094377289 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005348_120 |
0.1105691462 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_010080_070 |
0.1131152849 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 17 |
Hb_123689_010 |
0.1157991521 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 18 |
Hb_000390_160 |
0.1168577189 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 19 |
Hb_003905_030 |
0.1176932868 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 20 |
Hb_001268_170 |
0.1181246834 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |