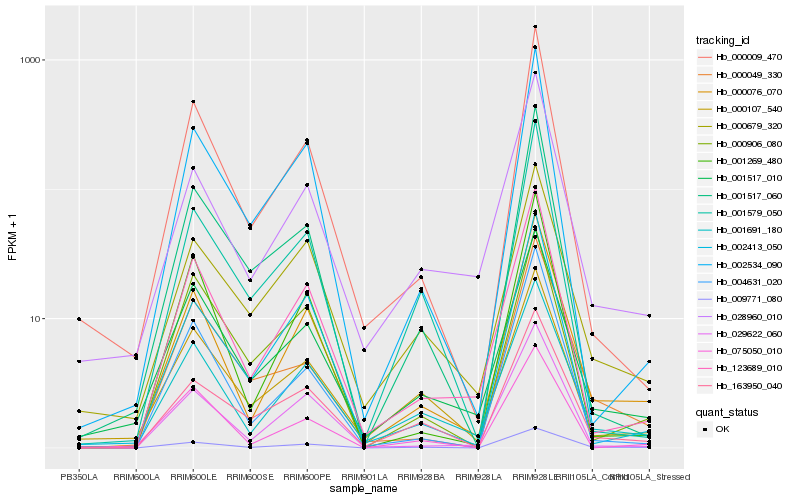
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009771_080 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 2 |
Hb_000076_070 |
0.0674403633 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 3 |
Hb_163950_040 |
0.0971625707 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001691_180 |
0.0973089122 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 5 |
Hb_123689_010 |
0.1295383328 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 6 |
Hb_000679_320 |
0.1297809141 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 7 |
Hb_001517_010 |
0.1334835411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002534_090 |
0.1341601138 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 9 |
Hb_000049_330 |
0.1369217431 |
- |
- |
PREDICTED: phospholipase A1-IIdelta [Jatropha curcas] |
| 10 |
Hb_000107_540 |
0.1371000667 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 11 |
Hb_028960_010 |
0.1388376572 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 12 |
Hb_000009_470 |
0.1389291751 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_001579_050 |
0.139890029 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_029622_060 |
0.1402565436 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002413_050 |
0.141579347 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 16 |
Hb_075050_010 |
0.1433373698 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001269_480 |
0.143534446 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004631_020 |
0.1436328611 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 19 |
Hb_001517_060 |
0.1444083224 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 20 |
Hb_000906_080 |
0.1460471579 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |