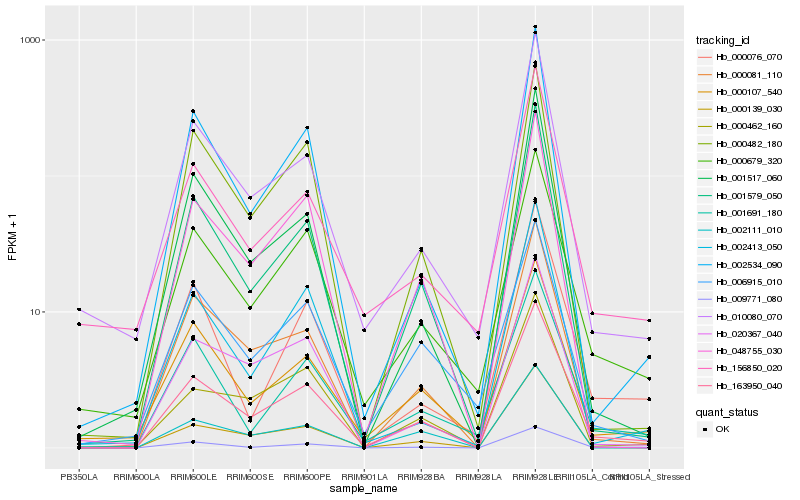
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163950_040 |
0.0 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_009771_080 |
0.0971625707 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 3 |
Hb_000081_110 |
0.0980250405 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_000679_320 |
0.0985185121 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 5 |
Hb_001579_050 |
0.1005908214 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000139_030 |
0.1011197272 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 7 |
Hb_048755_030 |
0.1092181429 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_000107_540 |
0.1116486104 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 9 |
Hb_010080_070 |
0.1140563726 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000462_160 |
0.118181026 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_020367_040 |
0.1194994206 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 12 |
Hb_001691_180 |
0.1203219789 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 13 |
Hb_002534_090 |
0.12115853 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 14 |
Hb_000482_180 |
0.1218937909 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 15 |
Hb_001517_060 |
0.1235726707 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 16 |
Hb_002111_010 |
0.1256433495 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 17 |
Hb_006915_010 |
0.1311152033 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_000076_070 |
0.1313160666 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 19 |
Hb_002413_050 |
0.1319909033 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 20 |
Hb_156850_020 |
0.1328532517 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |