| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
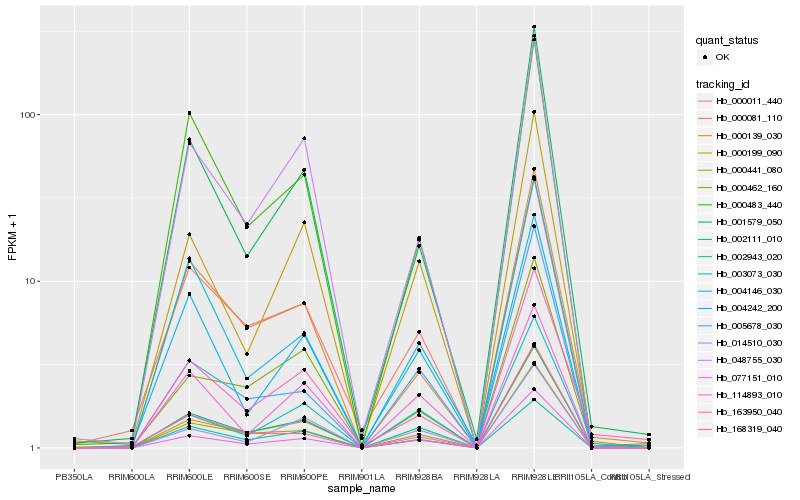
Hb_002111_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 2 |
Hb_000441_080 |
0.0425816304 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_077151_010 |
0.0645263868 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 4 |
Hb_000011_440 |
0.0909676139 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 5 |
Hb_048755_030 |
0.0910168979 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_001579_050 |
0.0972063884 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000139_030 |
0.0983379612 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 8 |
Hb_000199_090 |
0.0990068256 |
- |
- |
Tetrapyrrole-binding family protein [Populus trichocarpa] |
| 9 |
Hb_014510_030 |
0.1031406282 |
- |
- |
hypothetical protein VITISV_027576 [Vitis vinifera] |
| 10 |
Hb_003073_030 |
0.1043500964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000081_110 |
0.1069697695 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 12 |
Hb_168319_040 |
0.1115070039 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 13 |
Hb_000483_440 |
0.1137956213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002943_020 |
0.1151563053 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 15 |
Hb_004242_200 |
0.1180263357 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 16 |
Hb_114893_010 |
0.1187703034 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 17 |
Hb_004146_030 |
0.1201308798 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000462_160 |
0.1212564663 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_005678_030 |
0.122069017 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 20 |
Hb_163950_040 |
0.1256433495 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |