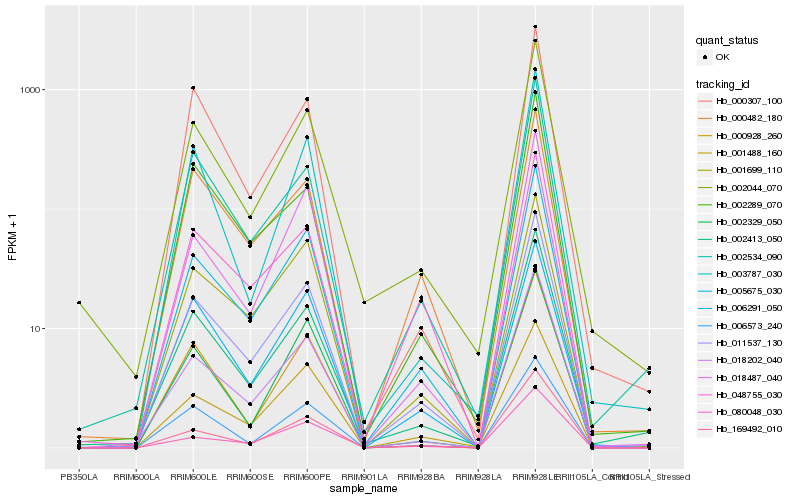
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011537_130 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_005675_030 |
0.0333473852 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 3 |
Hb_018202_040 |
0.0568467712 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_002413_050 |
0.058649457 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 5 |
Hb_002534_090 |
0.0716794706 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 6 |
Hb_169492_010 |
0.0755936668 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 7 |
Hb_002044_070 |
0.076419891 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 8 |
Hb_001488_160 |
0.0824568884 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_006573_240 |
0.0857525764 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 10 |
Hb_080048_030 |
0.0886655417 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 11 |
Hb_000928_260 |
0.0886788665 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 12 |
Hb_002289_070 |
0.090216277 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 13 |
Hb_048755_030 |
0.0906794003 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_018487_040 |
0.0925211117 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 15 |
Hb_001699_110 |
0.0944420252 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 16 |
Hb_000307_100 |
0.0946854432 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_003787_030 |
0.0965508677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000482_180 |
0.0969704757 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 19 |
Hb_002329_050 |
0.0969967487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006291_050 |
0.0974932482 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |