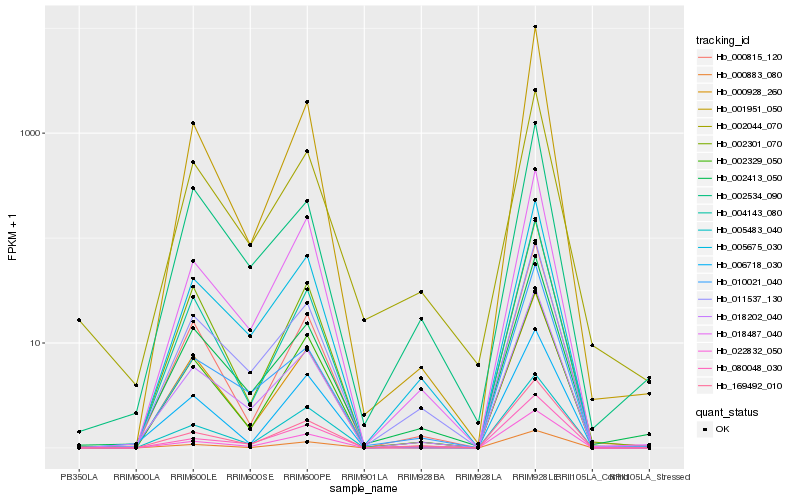
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018202_040 |
0.0 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_011537_130 |
0.0568467712 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_005675_030 |
0.0640352055 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 4 |
Hb_169492_010 |
0.0696304886 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 5 |
Hb_002413_050 |
0.0718733349 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 6 |
Hb_010021_040 |
0.0801935502 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 7 |
Hb_000883_080 |
0.0831623455 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_018487_040 |
0.0843819719 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 9 |
Hb_080048_030 |
0.0864721304 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 10 |
Hb_000928_260 |
0.0875308614 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 11 |
Hb_004143_080 |
0.0915454806 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 12 |
Hb_005483_040 |
0.0930286994 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 13 |
Hb_000815_120 |
0.0940258251 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_002044_070 |
0.0957048547 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 15 |
Hb_001951_050 |
0.0965628577 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 16 |
Hb_002534_090 |
0.0969286296 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 17 |
Hb_002329_050 |
0.098086881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002301_070 |
0.1003192337 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_006718_030 |
0.1006567515 |
- |
- |
ATPase, F1 complex, gamma subunit protein [Theobroma cacao] |
| 20 |
Hb_022832_050 |
0.101213419 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Sesamum indicum] |