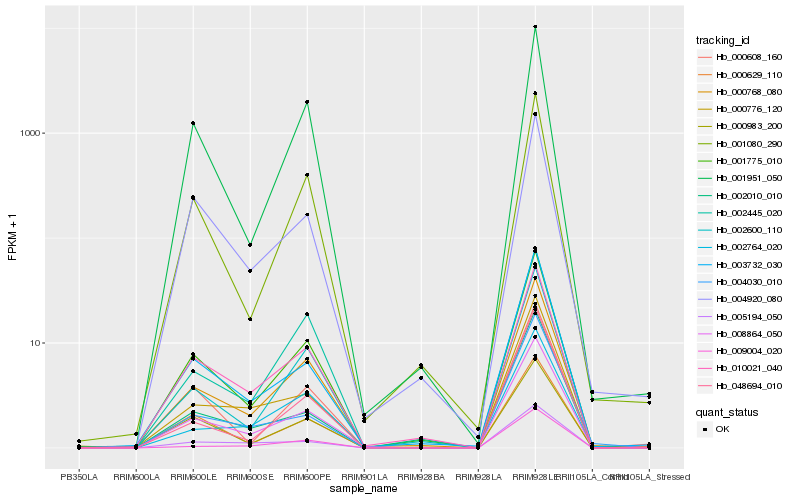
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008864_050 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_001775_010 |
0.0583267302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000768_080 |
0.0598538651 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 4 |
Hb_010021_040 |
0.06214056 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 5 |
Hb_003732_030 |
0.0808397077 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 6 |
Hb_000776_120 |
0.0835457755 |
- |
- |
PREDICTED: cytochrome P450 78A5 [Jatropha curcas] |
| 7 |
Hb_004920_080 |
0.0925102792 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_005194_050 |
0.0952872774 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 9 |
Hb_002600_110 |
0.0992609637 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 10 |
Hb_001080_290 |
0.0992656849 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 11 |
Hb_000629_110 |
0.1002637587 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 12 |
Hb_001951_050 |
0.104081103 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 13 |
Hb_002764_020 |
0.1048742715 |
- |
- |
hypothetical protein JCGZ_24084 [Jatropha curcas] |
| 14 |
Hb_004030_010 |
0.1060302442 |
- |
- |
hypothetical protein CICLE_v10013050mg [Citrus clementina] |
| 15 |
Hb_002445_020 |
0.1097531789 |
- |
- |
- |
| 16 |
Hb_000608_160 |
0.1097919631 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 17 |
Hb_048694_010 |
0.109990392 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_009004_020 |
0.1113069723 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 19 |
Hb_002010_010 |
0.1120953936 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 20 |
Hb_000983_200 |
0.1124562291 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |