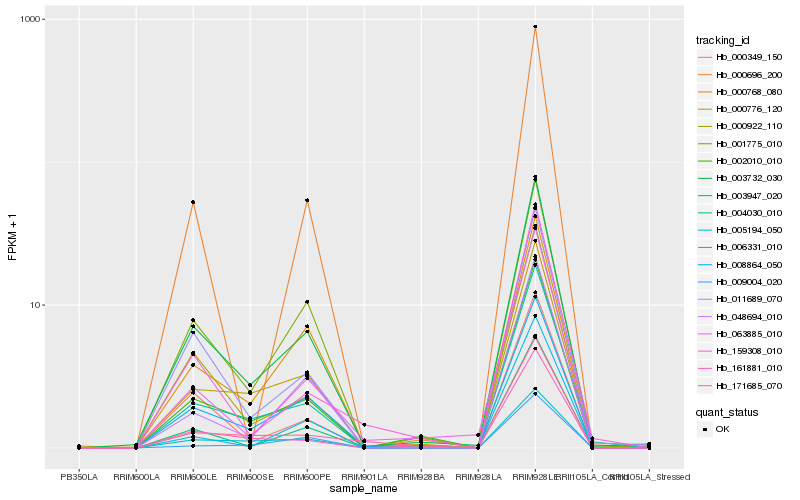
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004030_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10013050mg [Citrus clementina] |
| 2 |
Hb_003732_030 |
0.0713343895 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 3 |
Hb_000776_120 |
0.0775648348 |
- |
- |
PREDICTED: cytochrome P450 78A5 [Jatropha curcas] |
| 4 |
Hb_002010_010 |
0.0848888262 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 5 |
Hb_161881_010 |
0.0997755207 |
- |
- |
laccase, putative [Ricinus communis] |
| 6 |
Hb_011689_070 |
0.1002349174 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_008864_050 |
0.1060302442 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_000922_110 |
0.1076993643 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 9 |
Hb_001775_010 |
0.1139519848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005194_050 |
0.1142655399 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 11 |
Hb_048694_010 |
0.1148346782 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_000768_080 |
0.1193129479 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 13 |
Hb_003947_020 |
0.1229214384 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 14 |
Hb_171685_070 |
0.126292691 |
- |
- |
- |
| 15 |
Hb_000349_150 |
0.1272383439 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 16 |
Hb_009004_020 |
0.1276071476 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 17 |
Hb_006331_010 |
0.1276087797 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 18 |
Hb_063885_010 |
0.1280256377 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_159308_010 |
0.1293724535 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 20 |
Hb_000696_200 |
0.1296164041 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |