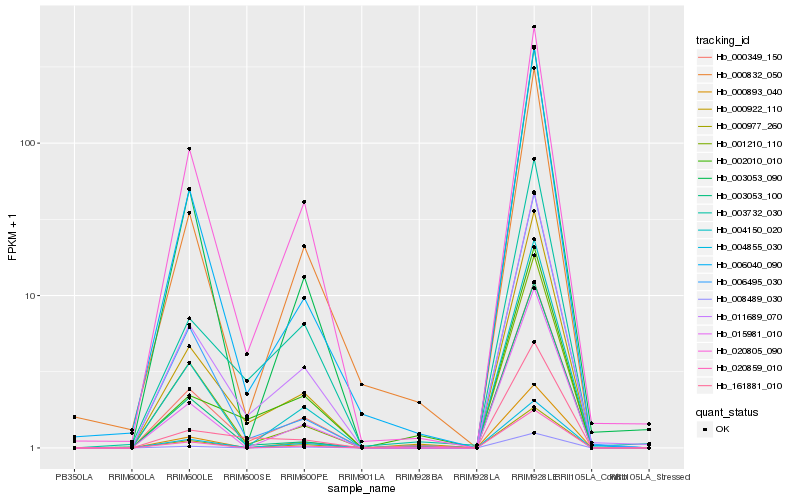
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_110 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 2 |
Hb_000349_150 |
0.0400211043 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 3 |
Hb_011689_070 |
0.0503171232 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004150_020 |
0.071761402 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 5 |
Hb_006040_090 |
0.0721548909 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Hevea brasiliensis] |
| 6 |
Hb_004855_030 |
0.0721836798 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 7 |
Hb_001210_110 |
0.0761913216 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 8 |
Hb_003732_030 |
0.0803560019 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 9 |
Hb_015981_010 |
0.0808867769 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_020859_010 |
0.0820354942 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X6 [Populus euphratica] |
| 11 |
Hb_003053_090 |
0.0839650528 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_008489_030 |
0.0861996114 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 13 |
Hb_020805_090 |
0.0865248985 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000893_040 |
0.0871451916 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 15 |
Hb_006495_030 |
0.0873787059 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 16 |
Hb_003053_100 |
0.0883387965 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 17 |
Hb_161881_010 |
0.0894948733 |
- |
- |
laccase, putative [Ricinus communis] |
| 18 |
Hb_000832_050 |
0.0932826052 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Marila laxiflora] |
| 19 |
Hb_002010_010 |
0.0957631519 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 20 |
Hb_000977_260 |
0.0959028912 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |