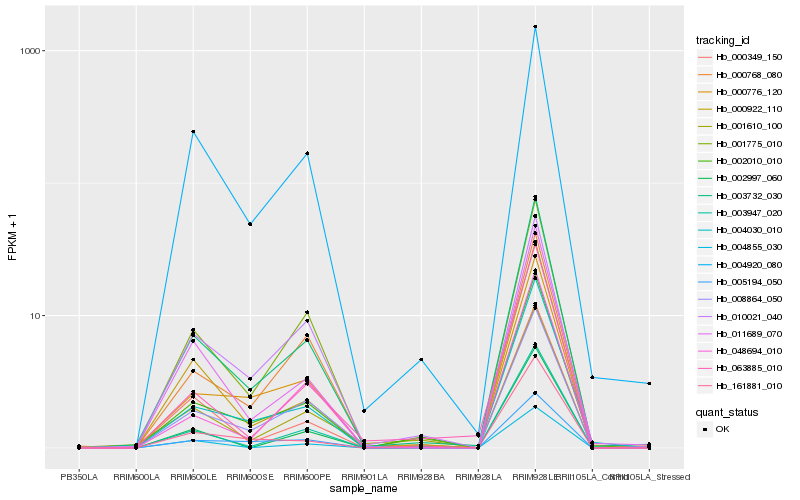
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003732_030 |
0.0 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 2 |
Hb_002010_010 |
0.0688955621 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 3 |
Hb_004030_010 |
0.0713343895 |
- |
- |
hypothetical protein CICLE_v10013050mg [Citrus clementina] |
| 4 |
Hb_011689_070 |
0.0717647307 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000776_120 |
0.0755868905 |
- |
- |
PREDICTED: cytochrome P450 78A5 [Jatropha curcas] |
| 6 |
Hb_001775_010 |
0.0776811241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000922_110 |
0.0803560019 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 8 |
Hb_008864_050 |
0.0808397077 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_003947_020 |
0.081305802 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 10 |
Hb_000349_150 |
0.0886439708 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 11 |
Hb_000768_080 |
0.0960166413 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 12 |
Hb_161881_010 |
0.0973577538 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_001610_100 |
0.0974108008 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_004855_030 |
0.0981400893 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 15 |
Hb_004920_080 |
0.1006326729 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_063885_010 |
0.1011830799 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_048694_010 |
0.1034979608 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_010021_040 |
0.1035684271 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 19 |
Hb_005194_050 |
0.1055463434 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 20 |
Hb_002997_060 |
0.1066806873 |
- |
- |
hypothetical protein POPTR_0011s10730g [Populus trichocarpa] |