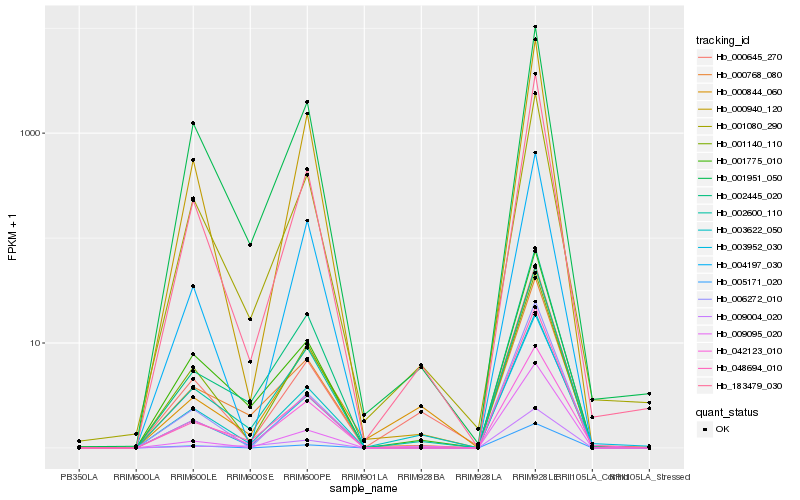
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002600_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 2 |
Hb_183479_030 |
0.0603780173 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000768_080 |
0.0645871185 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 4 |
Hb_048694_010 |
0.0673809642 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_001775_010 |
0.0706212626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001080_290 |
0.0715998364 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 7 |
Hb_002445_020 |
0.0741810774 |
- |
- |
- |
| 8 |
Hb_003622_050 |
0.0761447624 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |
| 9 |
Hb_003952_030 |
0.0776334715 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 10 |
Hb_000940_120 |
0.0799675673 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 11 |
Hb_001140_110 |
0.0879124762 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 12 |
Hb_004197_030 |
0.0917107561 |
- |
- |
hypothetical protein JCGZ_16263 [Jatropha curcas] |
| 13 |
Hb_001951_050 |
0.0922529925 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 14 |
Hb_009004_020 |
0.0924161294 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_006272_010 |
0.0942019627 |
- |
- |
PREDICTED: cytochrome P450 86B1 [Vitis vinifera] |
| 16 |
Hb_009095_020 |
0.0964331731 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 17 |
Hb_005171_020 |
0.0971567246 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 18 |
Hb_000844_060 |
0.097566062 |
- |
- |
PREDICTED: uncharacterized protein LOC105633706 [Jatropha curcas] |
| 19 |
Hb_042123_010 |
0.0975864456 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 20 |
Hb_000645_270 |
0.0982514454 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |