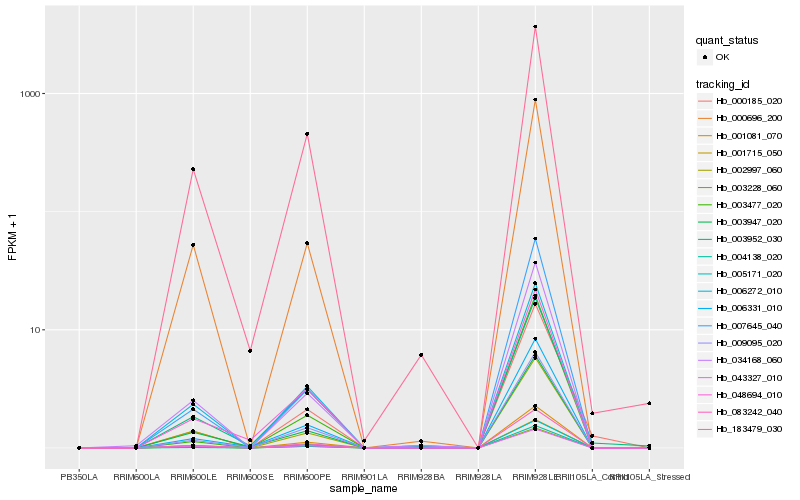
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006331_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 2 |
Hb_048694_010 |
0.0488751057 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_001081_070 |
0.0521113777 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 4 |
Hb_083242_040 |
0.0524812774 |
- |
- |
- |
| 5 |
Hb_004138_020 |
0.0557913628 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 6 |
Hb_001715_050 |
0.0577314334 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase AOP1 [Jatropha curcas] |
| 7 |
Hb_009095_020 |
0.0626053436 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 8 |
Hb_003228_060 |
0.0635546147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_034168_060 |
0.0705613295 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |
| 10 |
Hb_006272_010 |
0.0706194761 |
- |
- |
PREDICTED: cytochrome P450 86B1 [Vitis vinifera] |
| 11 |
Hb_000696_200 |
0.0751633618 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 12 |
Hb_007645_040 |
0.0809267725 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Tachigali myrmecophila] |
| 13 |
Hb_003477_020 |
0.0847533795 |
desease resistance |
Gene Name: AAA_29 |
PREDICTED: ABC transporter G family member 15-like [Populus euphratica] |
| 14 |
Hb_005171_020 |
0.0867498539 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 15 |
Hb_183479_030 |
0.0889112138 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_003952_030 |
0.0909514286 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 17 |
Hb_000185_020 |
0.0945513544 |
- |
- |
- |
| 18 |
Hb_043327_010 |
0.0958548322 |
- |
- |
PREDICTED: uncharacterized protein LOC105785093, partial [Gossypium raimondii] |
| 19 |
Hb_002997_060 |
0.0996929313 |
- |
- |
hypothetical protein POPTR_0011s10730g [Populus trichocarpa] |
| 20 |
Hb_003947_020 |
0.0997779182 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |