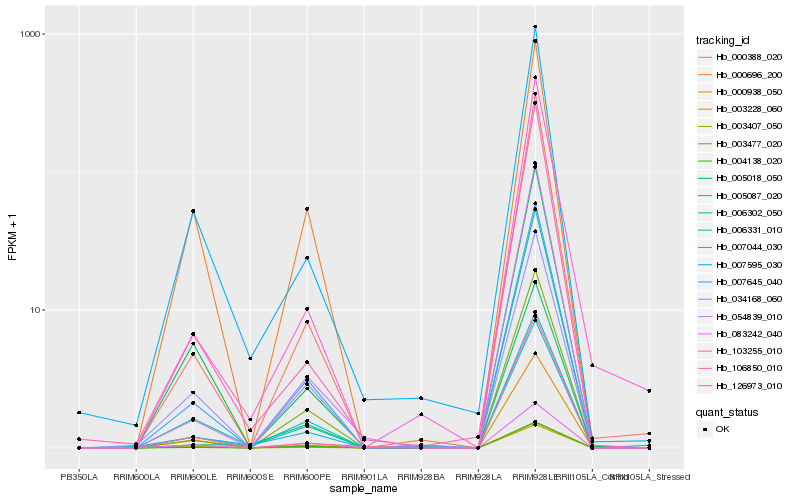
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007645_040 |
0.0 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Tachigali myrmecophila] |
| 2 |
Hb_004138_020 |
0.0468957723 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 3 |
Hb_003477_020 |
0.0538528061 |
desease resistance |
Gene Name: AAA_29 |
PREDICTED: ABC transporter G family member 15-like [Populus euphratica] |
| 4 |
Hb_000388_020 |
0.0596907759 |
- |
- |
PREDICTED: MLP-like protein 43 [Fragaria vesca subsp. vesca] |
| 5 |
Hb_034168_060 |
0.0654470727 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |
| 6 |
Hb_003407_050 |
0.067395331 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_054839_010 |
0.0757308246 |
- |
- |
PsbC, partial (chloroplast) [Asimina parviflora] |
| 8 |
Hb_005087_020 |
0.0793913179 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_006331_010 |
0.0809267725 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 10 |
Hb_005018_050 |
0.0819340077 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |
| 11 |
Hb_007044_030 |
0.0829017915 |
- |
- |
Os06g0273400 [Oryza sativa Japonica Group] |
| 12 |
Hb_106850_010 |
0.0839332467 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 13 |
Hb_006302_050 |
0.0864807127 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 14 |
Hb_126973_010 |
0.0875190799 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |
| 15 |
Hb_103255_010 |
0.0878888892 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 16 |
Hb_083242_040 |
0.0891605042 |
- |
- |
- |
| 17 |
Hb_007595_030 |
0.089547155 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 18 |
Hb_003228_060 |
0.0896102857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000696_200 |
0.0903555566 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 20 |
Hb_000938_050 |
0.0913265934 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |