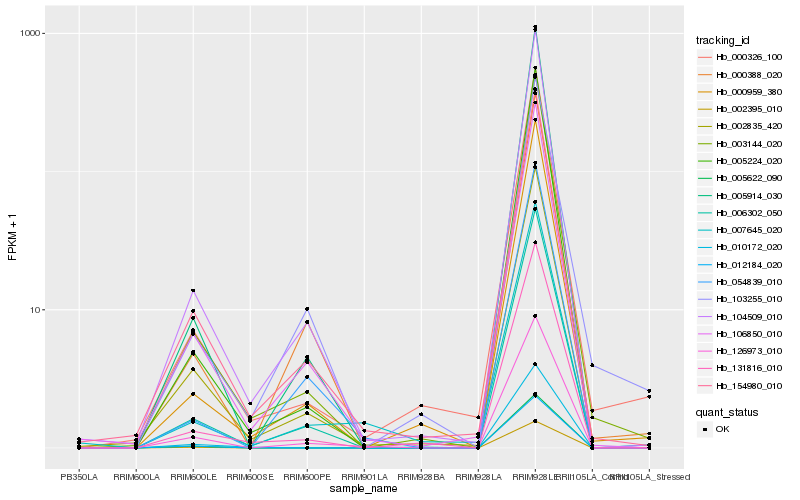
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006302_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 2 |
Hb_104509_010 |
0.0164406443 |
- |
- |
photosystem II CP43 chlorophyll apoprotein (chloroplast) [Ficus sp. Moore 315] |
| 3 |
Hb_106850_010 |
0.0375867388 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 4 |
Hb_005914_030 |
0.0421015285 |
- |
- |
- |
| 5 |
Hb_003144_020 |
0.0432435813 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 6 |
Hb_131816_010 |
0.0447963887 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 7 |
Hb_000959_380 |
0.0476718564 |
- |
- |
PREDICTED: (-)-alpha-terpineol synthase-like [Vitis vinifera] |
| 8 |
Hb_005224_020 |
0.0477836117 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_154980_010 |
0.0513709396 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 10 |
Hb_000388_020 |
0.0521933058 |
- |
- |
PREDICTED: MLP-like protein 43 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_002835_420 |
0.0524541577 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 12 |
Hb_126973_010 |
0.0602049511 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |
| 13 |
Hb_054839_010 |
0.0624318434 |
- |
- |
PsbC, partial (chloroplast) [Asimina parviflora] |
| 14 |
Hb_000326_100 |
0.0694699983 |
transcription factor |
TF Family: MYB-related |
DnaJ homolog subfamily C member 2 [Morus notabilis] |
| 15 |
Hb_005622_090 |
0.0729244662 |
- |
- |
hypothetical protein CICLE_v10027074mg [Citrus clementina] |
| 16 |
Hb_002395_010 |
0.0737980931 |
- |
- |
- |
| 17 |
Hb_012184_020 |
0.0756940969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010172_020 |
0.0780146797 |
- |
- |
PREDICTED: tyrosine/DOPA decarboxylase 1-like [Jatropha curcas] |
| 19 |
Hb_103255_010 |
0.0805413409 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 20 |
Hb_007645_020 |
0.0814855453 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit [Medicago truncatula] |