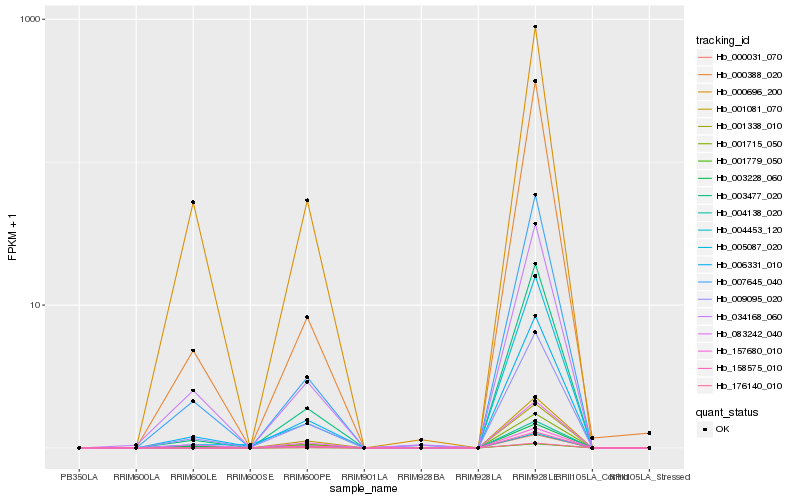
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004138_020 |
0.0 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 2 |
Hb_003477_020 |
0.0346511552 |
desease resistance |
Gene Name: AAA_29 |
PREDICTED: ABC transporter G family member 15-like [Populus euphratica] |
| 3 |
Hb_007645_040 |
0.0468957723 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Tachigali myrmecophila] |
| 4 |
Hb_001081_070 |
0.0506952423 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 5 |
Hb_006331_010 |
0.0557913628 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 6 |
Hb_034168_060 |
0.0758352386 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |
| 7 |
Hb_083242_040 |
0.0763595085 |
- |
- |
- |
| 8 |
Hb_005087_020 |
0.0819201942 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_001338_010 |
0.0835857224 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 10 |
Hb_000031_070 |
0.0837884738 |
- |
- |
Thioredoxin domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001779_050 |
0.0843398601 |
- |
- |
polyprotein [Ananas comosus] |
| 12 |
Hb_176140_010 |
0.0844683263 |
- |
- |
polyprotein [Oryza australiensis] |
| 13 |
Hb_003228_060 |
0.0847332964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001715_050 |
0.085415297 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase AOP1 [Jatropha curcas] |
| 15 |
Hb_000388_020 |
0.0861381239 |
- |
- |
PREDICTED: MLP-like protein 43 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_009095_020 |
0.087437444 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 17 |
Hb_158575_010 |
0.0912339276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_157680_010 |
0.0914742004 |
- |
- |
PREDICTED: uncharacterized protein LOC103444007 [Malus domestica] |
| 19 |
Hb_004453_120 |
0.0926693879 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 20 |
Hb_000696_200 |
0.0935131824 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |