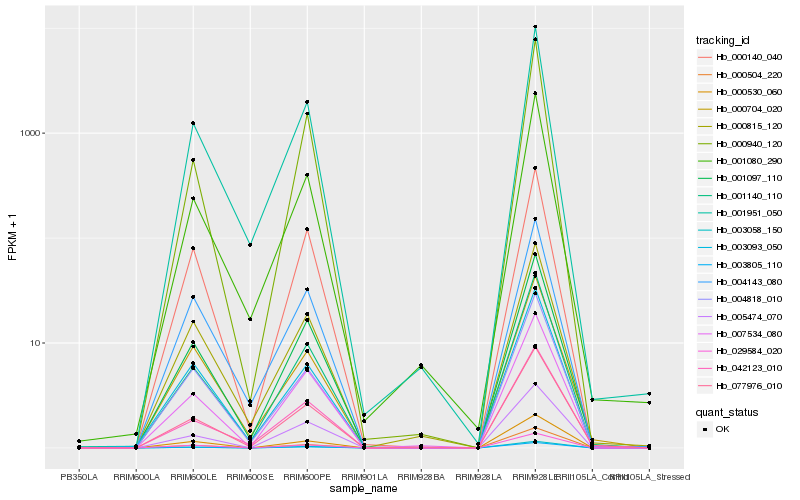
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077976_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 2 |
Hb_042123_010 |
0.0454110225 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 3 |
Hb_001097_110 |
0.0534033167 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 4 |
Hb_001140_110 |
0.0564854953 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 5 |
Hb_001080_290 |
0.0578301428 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 6 |
Hb_001951_050 |
0.0587850123 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 7 |
Hb_000815_120 |
0.0667917169 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_007534_080 |
0.068093558 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_003058_150 |
0.0708359541 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 10 |
Hb_003805_110 |
0.0714380036 |
- |
- |
PREDICTED: laccase-11-like [Jatropha curcas] |
| 11 |
Hb_004818_010 |
0.0720238793 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 12 |
Hb_005474_070 |
0.0794870791 |
- |
- |
hypothetical protein JCGZ_10579 [Jatropha curcas] |
| 13 |
Hb_029584_020 |
0.0820193367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000940_120 |
0.0829444612 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 15 |
Hb_000704_020 |
0.083349461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000530_060 |
0.0835133967 |
- |
- |
- |
| 17 |
Hb_004143_080 |
0.0840542957 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 18 |
Hb_000140_040 |
0.0863966632 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 19 |
Hb_000504_220 |
0.086770053 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 20 |
Hb_003093_050 |
0.0869485492 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |