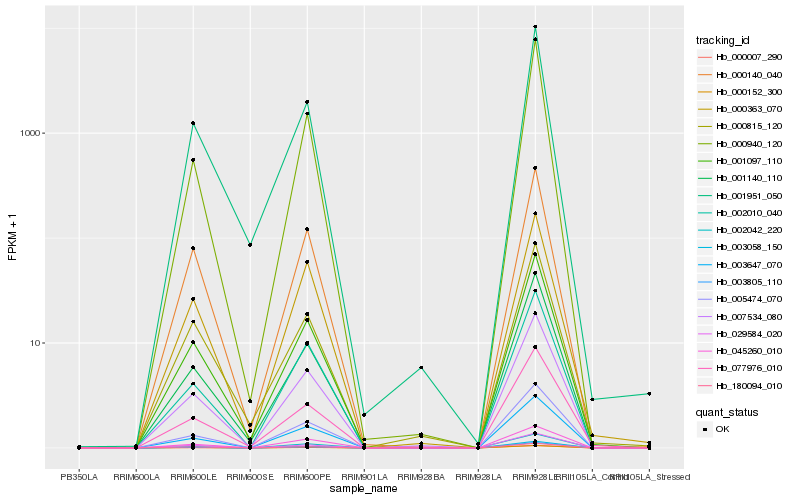
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007534_080 |
0.0 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_005474_070 |
0.0431335609 |
- |
- |
hypothetical protein JCGZ_10579 [Jatropha curcas] |
| 3 |
Hb_003647_070 |
0.0458954849 |
- |
- |
hypothetical protein POPTR_0006s09430g [Populus trichocarpa] |
| 4 |
Hb_003058_150 |
0.0486573479 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 5 |
Hb_001097_110 |
0.0501340981 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 6 |
Hb_000140_040 |
0.0515248167 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 7 |
Hb_003805_110 |
0.0529943046 |
- |
- |
PREDICTED: laccase-11-like [Jatropha curcas] |
| 8 |
Hb_001140_110 |
0.0545243226 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 9 |
Hb_002010_040 |
0.0554279994 |
- |
- |
- |
| 10 |
Hb_000007_290 |
0.0583302663 |
- |
- |
hypothetical protein AALP_AAs43195U000200 [Arabis alpina] |
| 11 |
Hb_045260_010 |
0.0664238148 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 12 |
Hb_000363_070 |
0.0670993652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_077976_010 |
0.068093558 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 14 |
Hb_001951_050 |
0.0684004797 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 15 |
Hb_002042_220 |
0.069648206 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 16 |
Hb_029584_020 |
0.0709379786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000815_120 |
0.07572644 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_180094_010 |
0.0760128145 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 19 |
Hb_000940_120 |
0.0761766921 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 20 |
Hb_000152_300 |
0.078072867 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |