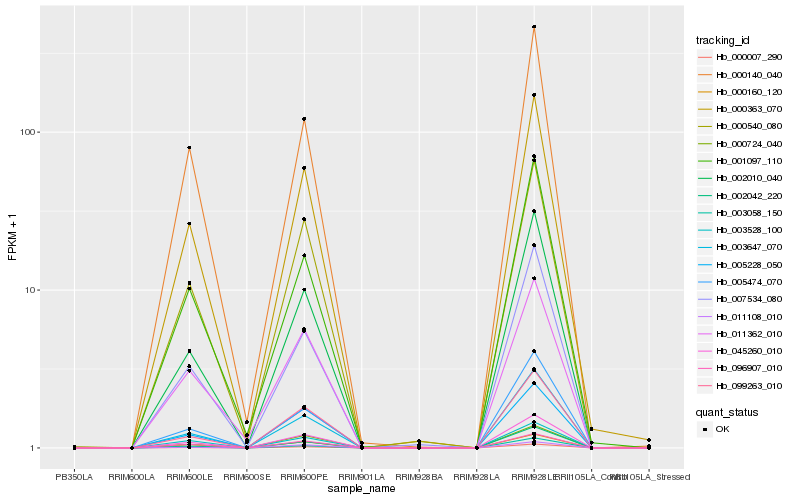
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_045260_010 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 2 |
Hb_000363_070 |
0.0381362986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003647_070 |
0.0386584261 |
- |
- |
hypothetical protein POPTR_0006s09430g [Populus trichocarpa] |
| 4 |
Hb_002010_040 |
0.0387500798 |
- |
- |
- |
| 5 |
Hb_000007_290 |
0.0415918965 |
- |
- |
hypothetical protein AALP_AAs43195U000200 [Arabis alpina] |
| 6 |
Hb_000540_080 |
0.0474884837 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 7 |
Hb_099263_010 |
0.0573507678 |
- |
- |
hypothetical protein AALP_AAs50029U000400 [Arabis alpina] |
| 8 |
Hb_005474_070 |
0.0580468695 |
- |
- |
hypothetical protein JCGZ_10579 [Jatropha curcas] |
| 9 |
Hb_000140_040 |
0.0652067622 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 10 |
Hb_007534_080 |
0.0664238148 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_005228_050 |
0.0686339937 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_000724_040 |
0.0742223616 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 13 |
Hb_000160_120 |
0.0743943315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002042_220 |
0.074962077 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 15 |
Hb_011362_010 |
0.0797763207 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 16 |
Hb_096907_010 |
0.0798025989 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 17 |
Hb_011108_010 |
0.0810043479 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103335582 [Prunus mume] |
| 18 |
Hb_003528_100 |
0.0820876513 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001097_110 |
0.0821882027 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 20 |
Hb_003058_150 |
0.0827706773 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |