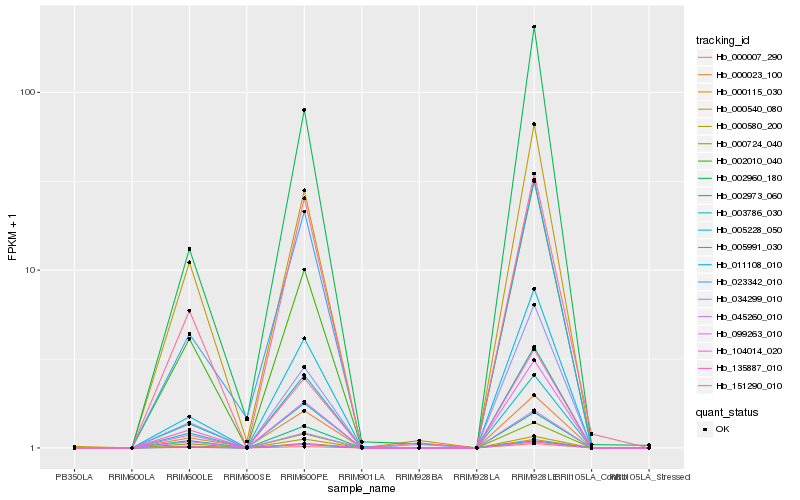
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011108_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103335582 [Prunus mume] |
| 2 |
Hb_135887_010 |
0.0276943287 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 3 |
Hb_005991_030 |
0.0312521961 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_005228_050 |
0.0338269344 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_000724_040 |
0.0382665472 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 6 |
Hb_003786_030 |
0.0423773713 |
- |
- |
BnaUnng00830D [Brassica napus] |
| 7 |
Hb_099263_010 |
0.0452465505 |
- |
- |
hypothetical protein AALP_AAs50029U000400 [Arabis alpina] |
| 8 |
Hb_002973_060 |
0.0616273353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000540_080 |
0.0725629527 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 10 |
Hb_000023_100 |
0.0764932259 |
- |
- |
Mechanosensitive channel of small conductance-like 10 [Theobroma cacao] |
| 11 |
Hb_045260_010 |
0.0810043479 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 12 |
Hb_000580_200 |
0.0835112267 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_023342_010 |
0.0860858291 |
- |
- |
- |
| 14 |
Hb_000115_030 |
0.0873657766 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 15 |
Hb_034299_010 |
0.0899653376 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 16 |
Hb_151290_010 |
0.0905400513 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_002960_180 |
0.091905021 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 18 |
Hb_104014_020 |
0.0920068065 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000007_290 |
0.0931362425 |
- |
- |
hypothetical protein AALP_AAs43195U000200 [Arabis alpina] |
| 20 |
Hb_002010_040 |
0.0946316353 |
- |
- |
- |