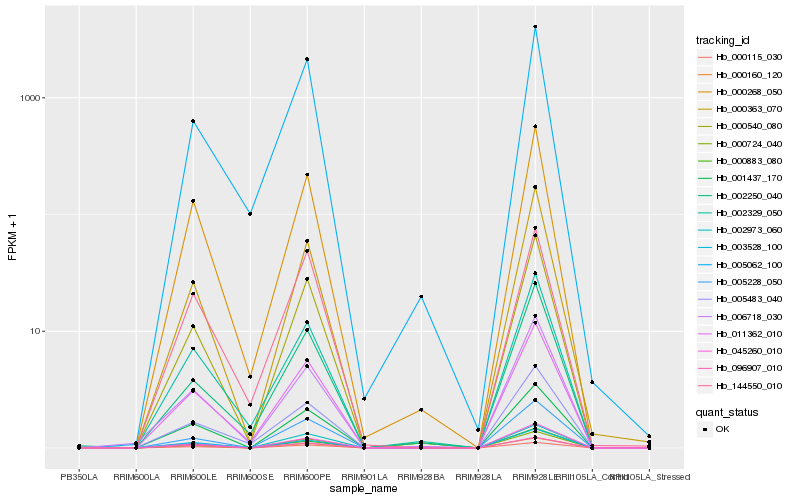
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011362_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 2 |
Hb_000268_050 |
0.0488631328 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002329_050 |
0.0539120949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000540_080 |
0.0570240226 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 5 |
Hb_000883_080 |
0.0626293274 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000160_120 |
0.0628726852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_096907_010 |
0.0634756692 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 8 |
Hb_001437_170 |
0.0658722817 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 9 |
Hb_000115_030 |
0.0701547122 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 10 |
Hb_005483_040 |
0.0734277112 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 11 |
Hb_005062_100 |
0.0742305277 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000363_070 |
0.0757317425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003528_100 |
0.0779011543 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 14 |
Hb_006718_030 |
0.0792566797 |
- |
- |
ATPase, F1 complex, gamma subunit protein [Theobroma cacao] |
| 15 |
Hb_002250_040 |
0.0796371971 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 16 |
Hb_045260_010 |
0.0797763207 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 17 |
Hb_144550_010 |
0.0808670497 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_002973_060 |
0.0814672421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005228_050 |
0.0817101024 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_000724_040 |
0.0818265749 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |