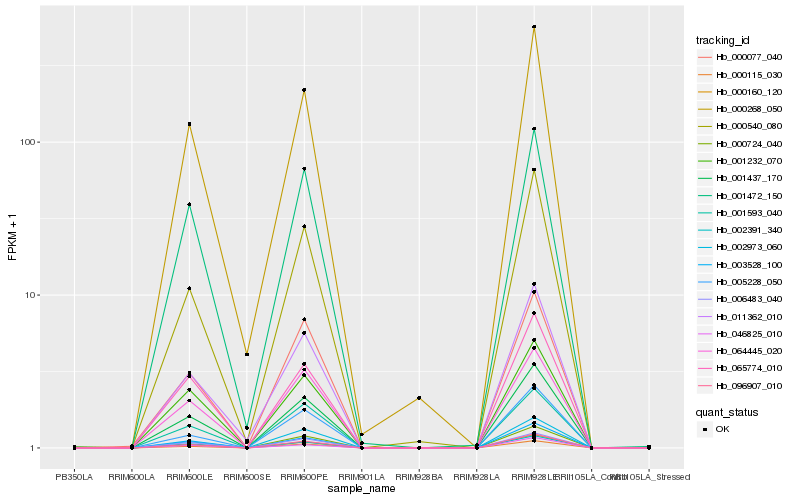
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096907_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 2 |
Hb_001437_170 |
0.0069182231 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 3 |
Hb_000160_120 |
0.0106435372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000115_030 |
0.0352237998 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 5 |
Hb_003528_100 |
0.044901319 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 6 |
Hb_002391_340 |
0.046979665 |
- |
- |
RNA polymerase II largest subunit [Liriodendron tulipifera] |
| 7 |
Hb_065774_010 |
0.0533664027 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000268_050 |
0.0582659309 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001472_150 |
0.0594568301 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 10 |
Hb_001232_070 |
0.0595008339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000540_080 |
0.0626280019 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 12 |
Hb_011362_010 |
0.0634756692 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 13 |
Hb_002973_060 |
0.064053938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006483_040 |
0.065942859 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 15 |
Hb_064445_020 |
0.0674721331 |
- |
- |
PREDICTED: serine carboxypeptidase-like 34 [Populus euphratica] |
| 16 |
Hb_000077_040 |
0.0682490664 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 17 |
Hb_001593_040 |
0.0684861453 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_000724_040 |
0.0737078252 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 19 |
Hb_005228_050 |
0.0756896645 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_046825_010 |
0.0781403229 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |