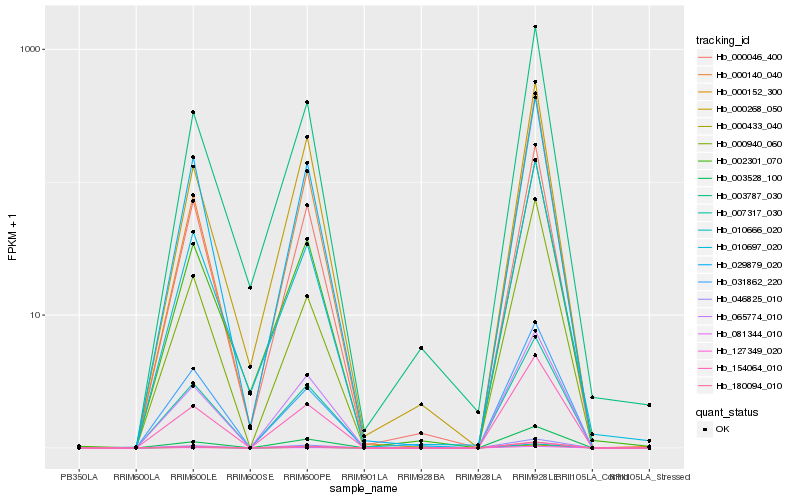
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_154064_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 2 |
Hb_046825_010 |
0.0245528125 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 3 |
Hb_127349_020 |
0.0342657894 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 4 |
Hb_180094_010 |
0.0401487965 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 5 |
Hb_003528_100 |
0.0465377442 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 6 |
Hb_007317_030 |
0.0470600615 |
- |
- |
- |
| 7 |
Hb_029879_020 |
0.048991815 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 8 |
Hb_065774_010 |
0.0494865882 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_010666_020 |
0.0553427932 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 10 |
Hb_000152_300 |
0.0586955369 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000433_040 |
0.0653922316 |
- |
- |
- |
| 12 |
Hb_000046_400 |
0.0662799195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_081344_010 |
0.0682359622 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 14 |
Hb_000140_040 |
0.0708394389 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 15 |
Hb_031862_220 |
0.0715791043 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |
| 16 |
Hb_000940_060 |
0.0732476371 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 17 |
Hb_002301_070 |
0.0738958873 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_003787_030 |
0.0772971573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010697_020 |
0.0788181905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000268_050 |
0.0790445244 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |