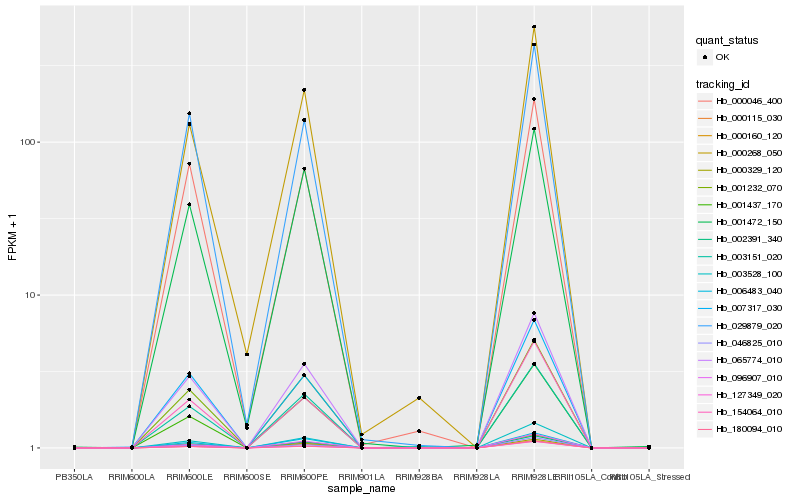
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065774_010 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003528_100 |
0.0252539409 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002391_340 |
0.0347935089 |
- |
- |
RNA polymerase II largest subunit [Liriodendron tulipifera] |
| 4 |
Hb_007317_030 |
0.0428743422 |
- |
- |
- |
| 5 |
Hb_001232_070 |
0.0445658212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001437_170 |
0.0468517139 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 7 |
Hb_000160_120 |
0.0470704874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_154064_010 |
0.0494865882 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 9 |
Hb_046825_010 |
0.0509818815 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 10 |
Hb_096907_010 |
0.0533664027 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 11 |
Hb_029879_020 |
0.0542294263 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 12 |
Hb_000046_400 |
0.0594687003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000268_050 |
0.0617483475 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001472_150 |
0.0705246162 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 15 |
Hb_000329_120 |
0.0728266639 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 16 |
Hb_003151_020 |
0.0742348966 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 17 |
Hb_127349_020 |
0.0751000941 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 18 |
Hb_180094_010 |
0.0759886769 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 19 |
Hb_006483_040 |
0.0776247965 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 20 |
Hb_000115_030 |
0.0874384697 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |