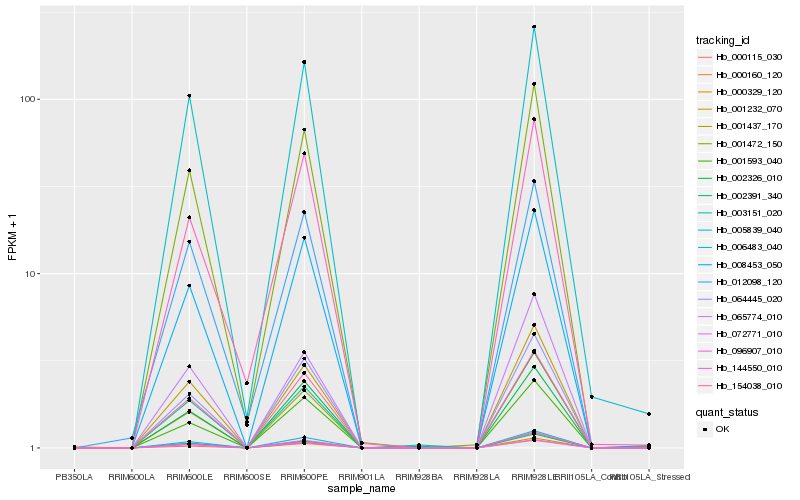
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001472_150 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 2 |
Hb_006483_040 |
0.0379463073 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 3 |
Hb_001232_070 |
0.0439443098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002391_340 |
0.0440395038 |
- |
- |
RNA polymerase II largest subunit [Liriodendron tulipifera] |
| 5 |
Hb_154038_010 |
0.0487499796 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_064445_020 |
0.0501400734 |
- |
- |
PREDICTED: serine carboxypeptidase-like 34 [Populus euphratica] |
| 7 |
Hb_008453_050 |
0.0555090362 |
- |
- |
PREDICTED: peroxidase 64 [Jatropha curcas] |
| 8 |
Hb_001437_170 |
0.0566330125 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 9 |
Hb_005839_040 |
0.0573141511 |
- |
- |
hypothetical protein POPTR_0019s08420g [Populus trichocarpa] |
| 10 |
Hb_001593_040 |
0.0584833125 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 11 |
Hb_096907_010 |
0.0594568301 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 12 |
Hb_003151_020 |
0.0613001182 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 13 |
Hb_000329_120 |
0.0633035765 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 14 |
Hb_000160_120 |
0.0649885647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_144550_010 |
0.0674482177 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_002326_010 |
0.0676066388 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 17 |
Hb_000115_030 |
0.0679067304 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 18 |
Hb_065774_010 |
0.0705246162 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_012098_120 |
0.0724561504 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 20 |
Hb_072771_010 |
0.0726141033 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |