| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
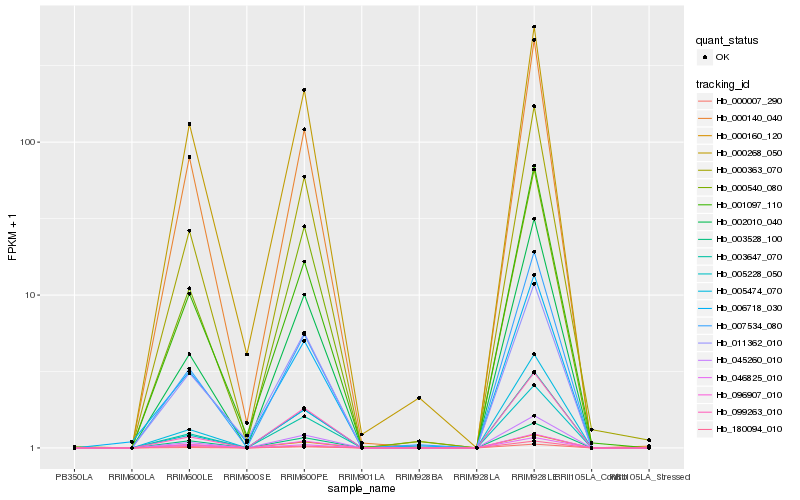
Hb_000363_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_045260_010 |
0.0381362986 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 3 |
Hb_000540_080 |
0.0481010904 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 4 |
Hb_003647_070 |
0.0573191254 |
- |
- |
hypothetical protein POPTR_0006s09430g [Populus trichocarpa] |
| 5 |
Hb_002010_040 |
0.0599185385 |
- |
- |
- |
| 6 |
Hb_000140_040 |
0.062483573 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 7 |
Hb_000007_290 |
0.0626769428 |
- |
- |
hypothetical protein AALP_AAs43195U000200 [Arabis alpina] |
| 8 |
Hb_007534_080 |
0.0670993652 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_005474_070 |
0.0722851173 |
- |
- |
hypothetical protein JCGZ_10579 [Jatropha curcas] |
| 10 |
Hb_000268_050 |
0.0741420467 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000160_120 |
0.0743682705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011362_010 |
0.0757317425 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 13 |
Hb_006718_030 |
0.0758624707 |
- |
- |
ATPase, F1 complex, gamma subunit protein [Theobroma cacao] |
| 14 |
Hb_001097_110 |
0.0765274774 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 15 |
Hb_099263_010 |
0.0780783454 |
- |
- |
hypothetical protein AALP_AAs50029U000400 [Arabis alpina] |
| 16 |
Hb_003528_100 |
0.0787560283 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 17 |
Hb_096907_010 |
0.080090292 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 18 |
Hb_046825_010 |
0.0807321345 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 19 |
Hb_180094_010 |
0.0807370989 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 20 |
Hb_005228_050 |
0.0813981404 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |