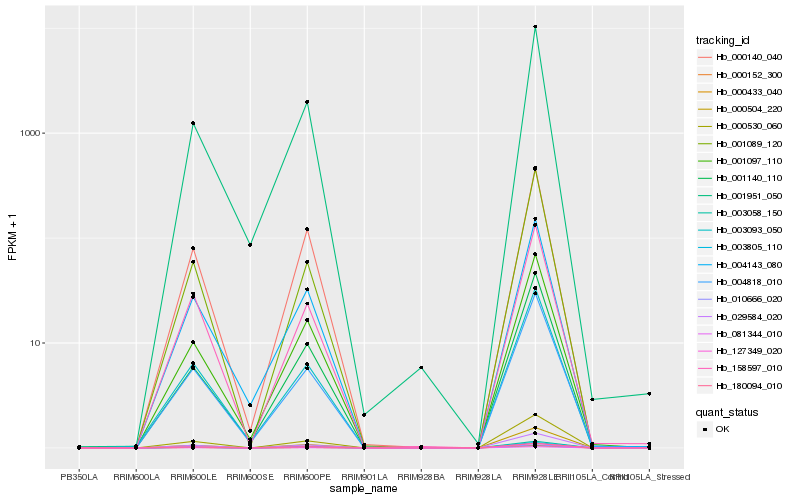
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029584_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000530_060 |
0.0221834736 |
- |
- |
- |
| 3 |
Hb_003805_110 |
0.0257760213 |
- |
- |
PREDICTED: laccase-11-like [Jatropha curcas] |
| 4 |
Hb_081344_010 |
0.0317911584 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 5 |
Hb_000433_040 |
0.0352012939 |
- |
- |
- |
| 6 |
Hb_000152_300 |
0.0391233989 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 7 |
Hb_010666_020 |
0.0460049391 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 8 |
Hb_003093_050 |
0.0476024308 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004818_010 |
0.0481305888 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 10 |
Hb_001089_120 |
0.0507749119 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 11 |
Hb_158597_010 |
0.0556249444 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000504_220 |
0.0588309287 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 13 |
Hb_001097_110 |
0.0589046429 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 14 |
Hb_000140_040 |
0.0593839134 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 15 |
Hb_001140_110 |
0.0624762277 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 16 |
Hb_180094_010 |
0.0629630082 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 17 |
Hb_003058_150 |
0.0635195978 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 18 |
Hb_127349_020 |
0.0646444438 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 19 |
Hb_004143_080 |
0.0666796572 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 20 |
Hb_001951_050 |
0.0700998185 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |