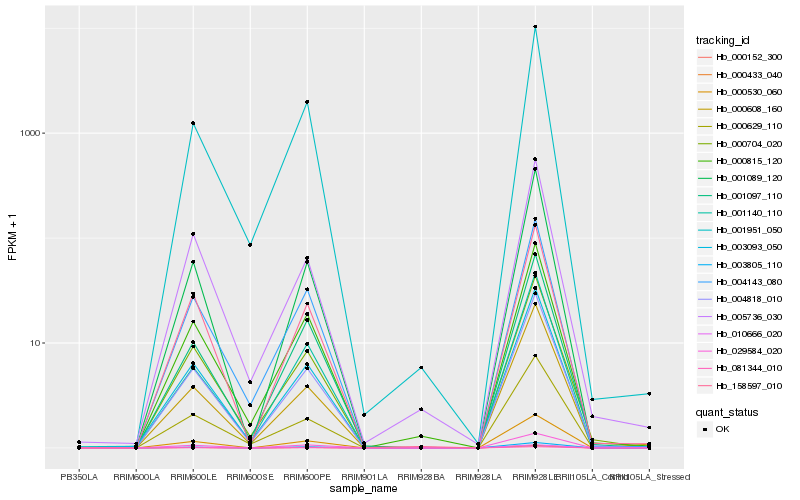
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003093_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004818_010 |
0.0317428351 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 3 |
Hb_029584_020 |
0.0476024308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000629_110 |
0.048247846 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 5 |
Hb_004143_080 |
0.0489237467 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 6 |
Hb_000530_060 |
0.0495543543 |
- |
- |
- |
| 7 |
Hb_000704_020 |
0.051778315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_081344_010 |
0.0552834218 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 9 |
Hb_000433_040 |
0.0570032779 |
- |
- |
- |
| 10 |
Hb_158597_010 |
0.0582621663 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000815_120 |
0.0586099576 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_003805_110 |
0.0610583466 |
- |
- |
PREDICTED: laccase-11-like [Jatropha curcas] |
| 13 |
Hb_000152_300 |
0.0620041584 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 14 |
Hb_001951_050 |
0.0621162561 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 15 |
Hb_001089_120 |
0.0633627747 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 16 |
Hb_010666_020 |
0.0640292971 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 17 |
Hb_000608_160 |
0.0659768031 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 18 |
Hb_001097_110 |
0.0674559912 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 19 |
Hb_001140_110 |
0.0693841082 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 20 |
Hb_005736_030 |
0.0735446113 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |