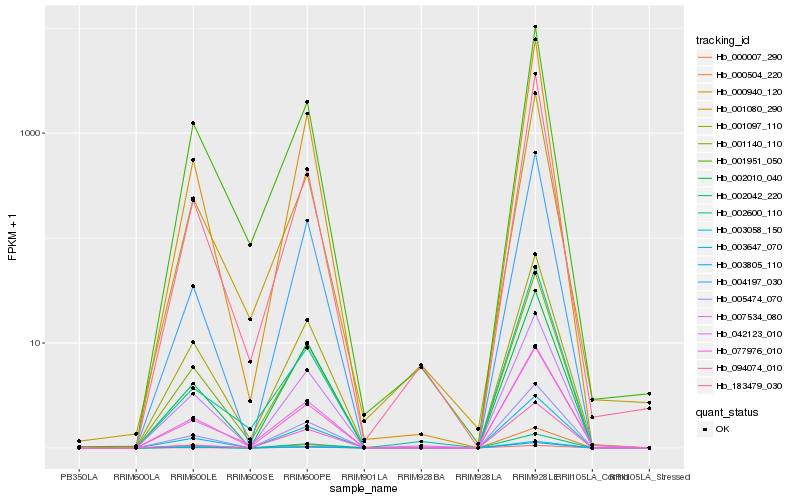
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000940_120 |
0.0 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 2 |
Hb_004197_030 |
0.0382297808 |
- |
- |
hypothetical protein JCGZ_16263 [Jatropha curcas] |
| 3 |
Hb_003058_150 |
0.0400428371 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 4 |
Hb_002042_220 |
0.0474985071 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 5 |
Hb_001140_110 |
0.0531059126 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 6 |
Hb_005474_070 |
0.0539071186 |
- |
- |
hypothetical protein JCGZ_10579 [Jatropha curcas] |
| 7 |
Hb_003647_070 |
0.0718489144 |
- |
- |
hypothetical protein POPTR_0006s09430g [Populus trichocarpa] |
| 8 |
Hb_000504_220 |
0.0723656838 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 9 |
Hb_003805_110 |
0.0731316803 |
- |
- |
PREDICTED: laccase-11-like [Jatropha curcas] |
| 10 |
Hb_000007_290 |
0.0732694977 |
- |
- |
hypothetical protein AALP_AAs43195U000200 [Arabis alpina] |
| 11 |
Hb_183479_030 |
0.0736371848 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_002010_040 |
0.0740364992 |
- |
- |
- |
| 13 |
Hb_001080_290 |
0.0754353726 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 14 |
Hb_007534_080 |
0.0761766921 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_001097_110 |
0.0788191957 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 16 |
Hb_001951_050 |
0.0790395207 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 17 |
Hb_002600_110 |
0.0799675673 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 18 |
Hb_042123_010 |
0.0814685704 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 19 |
Hb_077976_010 |
0.0829444612 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 20 |
Hb_094074_010 |
0.0899794557 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |