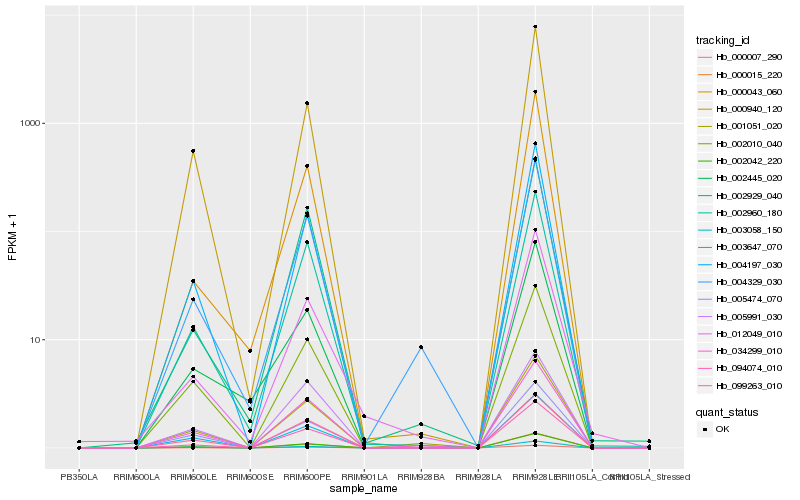
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094074_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 2 |
Hb_002960_180 |
0.0439297361 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 3 |
Hb_004197_030 |
0.0549660112 |
- |
- |
hypothetical protein JCGZ_16263 [Jatropha curcas] |
| 4 |
Hb_002042_220 |
0.0609115364 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 5 |
Hb_002929_040 |
0.0613601568 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_034299_010 |
0.077474978 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 7 |
Hb_099263_010 |
0.0788341178 |
- |
- |
hypothetical protein AALP_AAs50029U000400 [Arabis alpina] |
| 8 |
Hb_000015_220 |
0.081192668 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 9 |
Hb_000007_290 |
0.0816009859 |
- |
- |
hypothetical protein AALP_AAs43195U000200 [Arabis alpina] |
| 10 |
Hb_000043_060 |
0.0843074233 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 11 |
Hb_004329_030 |
0.0846668509 |
- |
- |
hypothetical protein JCGZ_17335 [Jatropha curcas] |
| 12 |
Hb_002010_040 |
0.0859051965 |
- |
- |
- |
| 13 |
Hb_005991_030 |
0.0891298735 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_000940_120 |
0.0899794557 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 15 |
Hb_001051_020 |
0.0947025329 |
- |
- |
hypothetical protein L484_008044 [Morus notabilis] |
| 16 |
Hb_005474_070 |
0.0960602595 |
- |
- |
hypothetical protein JCGZ_10579 [Jatropha curcas] |
| 17 |
Hb_003647_070 |
0.0971613987 |
- |
- |
hypothetical protein POPTR_0006s09430g [Populus trichocarpa] |
| 18 |
Hb_012049_010 |
0.1004683538 |
- |
- |
hypothetical protein B456_002G055100 [Gossypium raimondii] |
| 19 |
Hb_002445_020 |
0.1070867659 |
- |
- |
- |
| 20 |
Hb_003058_150 |
0.1083958331 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |