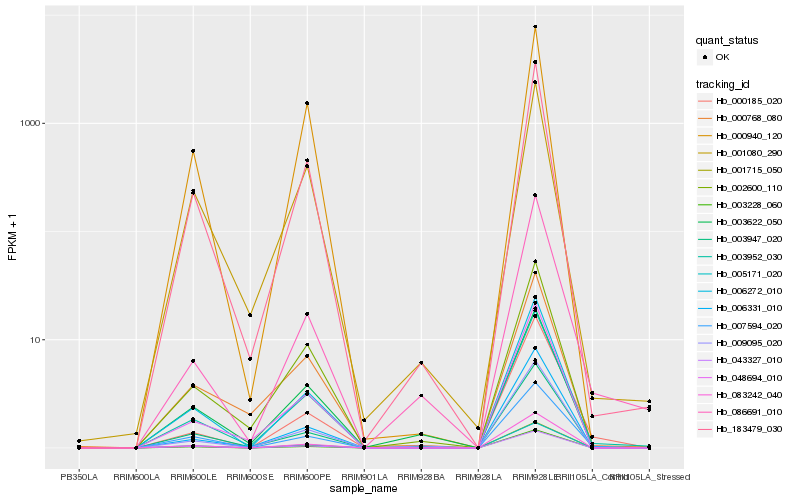
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003952_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 2 |
Hb_183479_030 |
0.0544306334 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_048694_010 |
0.0739108805 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_006272_010 |
0.0764396946 |
- |
- |
PREDICTED: cytochrome P450 86B1 [Vitis vinifera] |
| 5 |
Hb_002600_110 |
0.0776334715 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 6 |
Hb_005171_020 |
0.0821299681 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 7 |
Hb_001715_050 |
0.0847939831 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase AOP1 [Jatropha curcas] |
| 8 |
Hb_043327_010 |
0.0862156 |
- |
- |
PREDICTED: uncharacterized protein LOC105785093, partial [Gossypium raimondii] |
| 9 |
Hb_007594_020 |
0.0894439462 |
- |
- |
hypothetical protein JCGZ_18350 [Jatropha curcas] |
| 10 |
Hb_083242_040 |
0.0897474021 |
- |
- |
- |
| 11 |
Hb_009095_020 |
0.0904078553 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 12 |
Hb_003947_020 |
0.0906693426 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 13 |
Hb_006331_010 |
0.0909514286 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 14 |
Hb_001080_290 |
0.0942315043 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 15 |
Hb_086691_010 |
0.0961651535 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 16 |
Hb_000940_120 |
0.0981868857 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 17 |
Hb_003228_060 |
0.0983813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000768_080 |
0.0994506092 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 19 |
Hb_003622_050 |
0.1008239899 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A-like [Jatropha curcas] |
| 20 |
Hb_000185_020 |
0.1009803426 |
- |
- |
- |