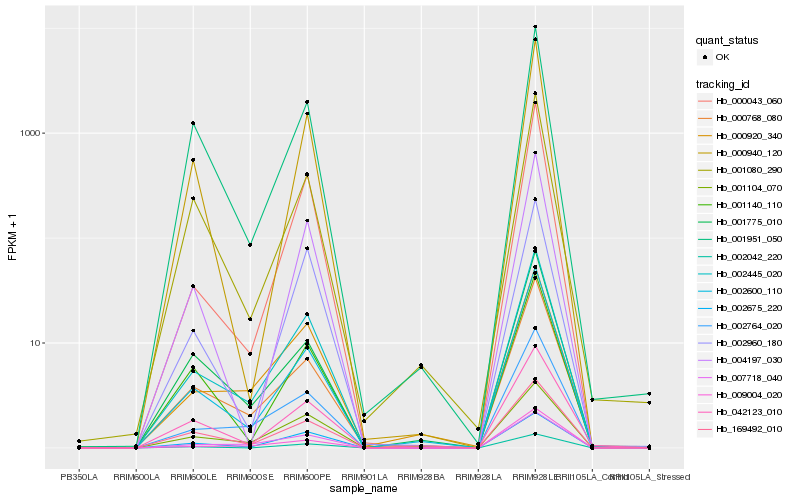
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002445_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002764_020 |
0.0682296096 |
- |
- |
hypothetical protein JCGZ_24084 [Jatropha curcas] |
| 3 |
Hb_002600_110 |
0.0741810774 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 4 |
Hb_000768_080 |
0.0790314914 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 5 |
Hb_000920_340 |
0.0853567994 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_042123_010 |
0.0866620245 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 7 |
Hb_007718_040 |
0.0869917211 |
- |
- |
hypothetical protein PRUPE_ppb025054mg [Prunus persica] |
| 8 |
Hb_004197_030 |
0.0909344718 |
- |
- |
hypothetical protein JCGZ_16263 [Jatropha curcas] |
| 9 |
Hb_000940_120 |
0.0931821462 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 10 |
Hb_001951_050 |
0.0967113254 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 11 |
Hb_002042_220 |
0.0969626648 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 12 |
Hb_001104_070 |
0.0973536538 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 13 |
Hb_000043_060 |
0.098482905 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 14 |
Hb_001140_110 |
0.0985494355 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 15 |
Hb_001080_290 |
0.0986158415 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 16 |
Hb_009004_020 |
0.0986619749 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 17 |
Hb_002960_180 |
0.102216792 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 18 |
Hb_169492_010 |
0.1023755706 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 19 |
Hb_002675_220 |
0.1036040654 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 20 |
Hb_001775_010 |
0.105111514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |