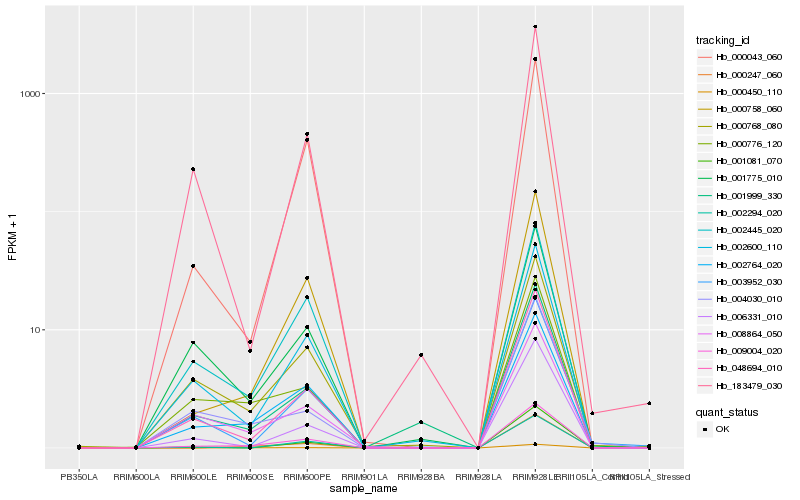
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009004_020 |
0.0 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_002764_020 |
0.0589903233 |
- |
- |
hypothetical protein JCGZ_24084 [Jatropha curcas] |
| 3 |
Hb_048694_010 |
0.0819904876 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000758_060 |
0.090224945 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45960 [Jatropha curcas] |
| 5 |
Hb_002600_110 |
0.0924161294 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 6 |
Hb_000768_080 |
0.0933146398 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 7 |
Hb_002445_020 |
0.0986619749 |
- |
- |
- |
| 8 |
Hb_000776_120 |
0.1017906572 |
- |
- |
PREDICTED: cytochrome P450 78A5 [Jatropha curcas] |
| 9 |
Hb_000247_060 |
0.108251096 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ORG2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_008864_050 |
0.1113069723 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_000043_060 |
0.1122901289 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 12 |
Hb_006331_010 |
0.1163965827 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 13 |
Hb_001775_010 |
0.1202770143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002294_020 |
0.1207067114 |
- |
- |
PREDICTED: uncharacterized protein LOC105629874 [Jatropha curcas] |
| 15 |
Hb_003952_030 |
0.1245617465 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 16 |
Hb_001999_330 |
0.1267232975 |
- |
- |
hypothetical protein PRUPE_ppa022262mg, partial [Prunus persica] |
| 17 |
Hb_001081_070 |
0.1273750148 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 18 |
Hb_004030_010 |
0.1276071476 |
- |
- |
hypothetical protein CICLE_v10013050mg [Citrus clementina] |
| 19 |
Hb_000450_110 |
0.1282620268 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 20 |
Hb_183479_030 |
0.1291426047 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |