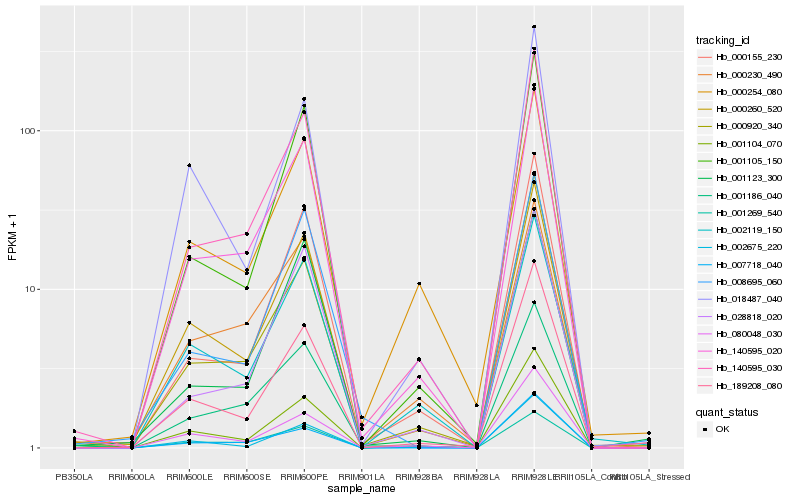
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140595_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_140595_020 |
0.0393590795 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |
| 3 |
Hb_000155_230 |
0.0659023931 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 4 |
Hb_001105_150 |
0.0715909696 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 5 |
Hb_001104_070 |
0.0718899621 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 6 |
Hb_000920_340 |
0.0723887162 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000260_520 |
0.0787413335 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_189208_080 |
0.0804438486 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001186_040 |
0.0816165235 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 10 |
Hb_028818_020 |
0.0848221851 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 11 |
Hb_007718_040 |
0.0876614151 |
- |
- |
hypothetical protein PRUPE_ppb025054mg [Prunus persica] |
| 12 |
Hb_080048_030 |
0.0947449761 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 13 |
Hb_002675_220 |
0.1037607223 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 14 |
Hb_001123_300 |
0.1123588933 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 15 |
Hb_008695_060 |
0.1135662135 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 16 |
Hb_002119_150 |
0.1138684302 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 17 |
Hb_018487_040 |
0.1157495078 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 18 |
Hb_000254_080 |
0.1210084527 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 19 |
Hb_001269_540 |
0.1219508759 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 20 |
Hb_000230_490 |
0.1238268984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |