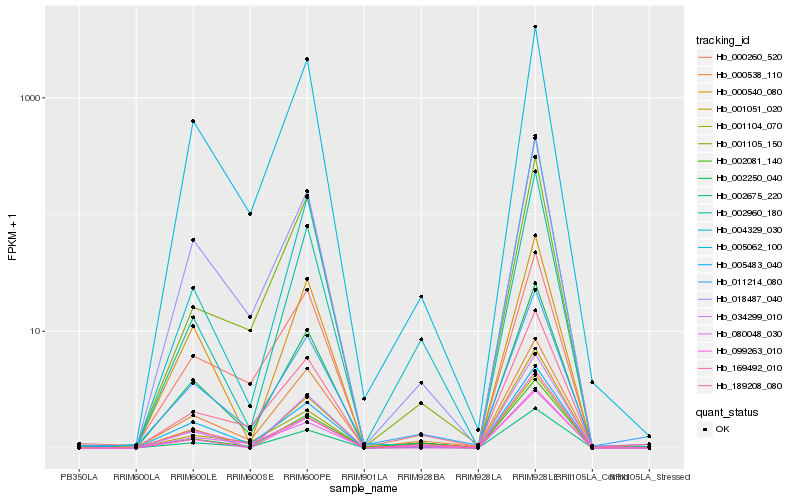
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002675_220 |
0.0 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 2 |
Hb_002250_040 |
0.0378503095 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 3 |
Hb_001104_070 |
0.0469735554 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 4 |
Hb_018487_040 |
0.0479543841 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 5 |
Hb_189208_080 |
0.0755142637 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_080048_030 |
0.0761374254 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 7 |
Hb_005483_040 |
0.0794878086 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 8 |
Hb_034299_010 |
0.08010126 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 9 |
Hb_001105_150 |
0.0840944607 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 10 |
Hb_169492_010 |
0.0860473198 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 11 |
Hb_005062_100 |
0.0865632893 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000260_520 |
0.0883315134 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_004329_030 |
0.0899344752 |
- |
- |
hypothetical protein JCGZ_17335 [Jatropha curcas] |
| 14 |
Hb_002960_180 |
0.0913078537 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 15 |
Hb_099263_010 |
0.0922368327 |
- |
- |
hypothetical protein AALP_AAs50029U000400 [Arabis alpina] |
| 16 |
Hb_000540_080 |
0.092705928 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 17 |
Hb_001051_020 |
0.0937445159 |
- |
- |
hypothetical protein L484_008044 [Morus notabilis] |
| 18 |
Hb_002081_140 |
0.0938180534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000538_110 |
0.0939911925 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 20 |
Hb_011214_080 |
0.0943272582 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |