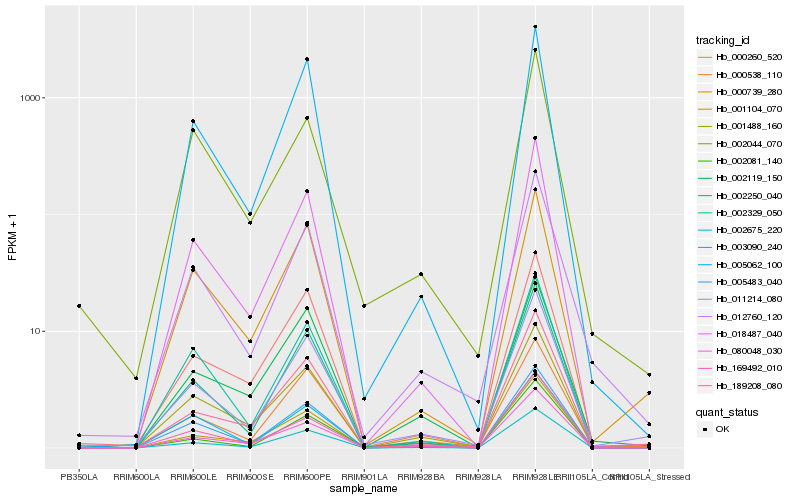
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011214_080 |
0.0 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 2 |
Hb_000538_110 |
0.0732579094 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 3 |
Hb_189208_080 |
0.0827631947 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_012760_120 |
0.0830297299 |
- |
- |
PREDICTED: peroxidase 64-like [Jatropha curcas] |
| 5 |
Hb_018487_040 |
0.0917743505 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 6 |
Hb_002250_040 |
0.0927900552 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 7 |
Hb_002675_220 |
0.0943272582 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 8 |
Hb_005483_040 |
0.0949294501 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 9 |
Hb_000739_280 |
0.0982392032 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_001488_160 |
0.0997604498 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_080048_030 |
0.1031343921 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 12 |
Hb_001104_070 |
0.1036598237 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 13 |
Hb_000260_520 |
0.1050955232 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_005062_100 |
0.1086293953 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_003090_240 |
0.1129496792 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 16 |
Hb_002329_050 |
0.1164116303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002081_140 |
0.1177450134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002044_070 |
0.1185828818 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 19 |
Hb_169492_010 |
0.1187345381 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 20 |
Hb_002119_150 |
0.1191089045 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |