Hb_005062_100
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig5062: 79680-80120 |
| Sequence |   |
Annotation
kegg
| ID | rcu:RCOM_0321060 |
|---|---|
| description | Ferredoxin-2, chloroplast precursor, putative |
nr
| ID | XP_002530673.1 |
|---|---|
| description | Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
swissprot
| ID | Q9ZTS2 |
|---|---|
| description | Ferredoxin, chloroplastic OS=Capsicum annuum GN=AP1 PE=1 SV=1 |
trembl
| ID | B9SXF4 |
|---|---|
| description | Ferredoxin-2, chloroplast, putative OS=Ricinus communis GN=RCOM_0321060 PE=4 SV=1 |
Gene Ontology
| ID | GO:0009055 |
|---|---|
| description | ferredoxin- chloroplast |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_46123: 79415-80193 |
|---|---|
| cDNA (Sanger) (ID:Location) |
002_J19.ab1: 79512-80164 , 005_B09.ab1: 79562-80164 , 006_A04.ab1: 79512-80164 , 014_B18.ab1: 79506-80164 , 021_I18.ab1: 79512-80164 , 022_O13.ab1: 79775-80164 , 025_G09.ab1: 79500-80164 , 026_K18.ab1: 79525-80164 , 029_H07.ab1: 79506-80164 , 030_H22.ab1: 79507-80164 , 032_C04.ab1: 79512-80164 , 034_L15.ab1: 79506-80164 , 037_I24.ab1: 79589-80164 , 038_G04.ab1: 79512-80193 , 038_P18.ab1: 79509-80164 , 039_O23.ab1: 79507-80164 , 043_H22.ab1: 79512-80164 , 048_A08.ab1: 79509-80164 , 052_H24.ab1: 79512-80174 , 053_F13.ab1: 79509-80164 , 053_H13.ab1: 79524-80193 , 053_I09.ab1: 79515-80164 |
Similar expressed genes (Top20)
Gene co-expression network
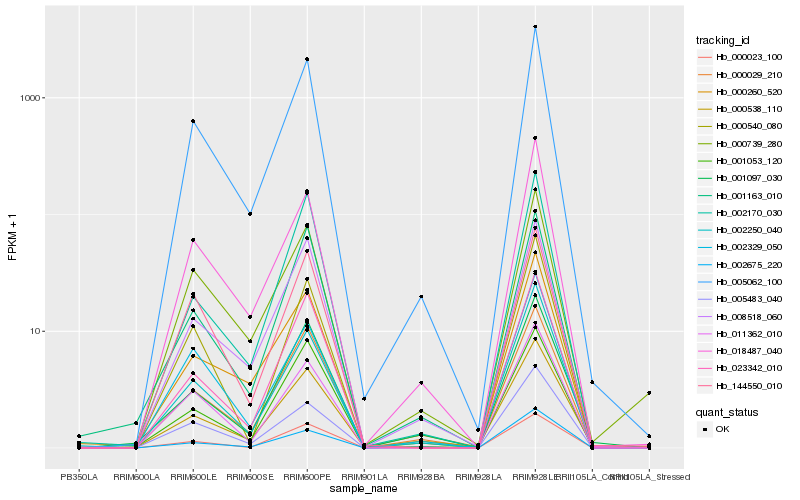
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 0.0705739 | 100.404 | 631.745 | 2140.78 | 0 | 1.64278 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 2.66182 | 0.261346 | 0.430068 | 18.8451 | 4079.67 |

