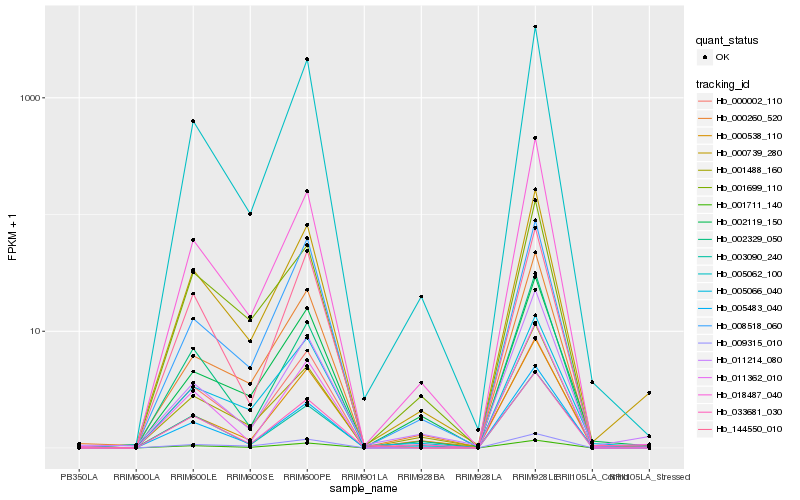
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_280 |
0.0 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_005062_100 |
0.0771159029 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_001488_160 |
0.0865908263 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_005066_040 |
0.0920555487 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 5 |
Hb_000002_110 |
0.0955917612 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 6 |
Hb_001699_110 |
0.0956336259 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 7 |
Hb_000260_520 |
0.0963314638 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000538_110 |
0.0973035117 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 9 |
Hb_018487_040 |
0.0973084543 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 10 |
Hb_001711_140 |
0.0982319304 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 11 |
Hb_011214_080 |
0.0982392032 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 12 |
Hb_003090_240 |
0.0983787608 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 13 |
Hb_002329_050 |
0.0985889659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005483_040 |
0.101442514 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 15 |
Hb_144550_010 |
0.1028080187 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_011362_010 |
0.1036215424 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 17 |
Hb_002119_150 |
0.1044146944 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 18 |
Hb_033681_030 |
0.1054347217 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 19 |
Hb_008518_060 |
0.1085847397 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 20 |
Hb_009315_010 |
0.1091457626 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |