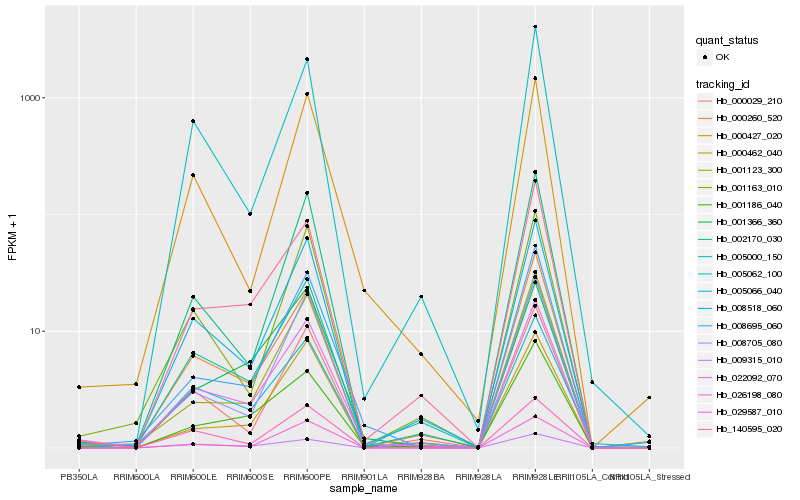
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022092_070 |
0.0 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.5 [Jatropha curcas] |
| 2 |
Hb_008518_060 |
0.0831204221 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 3 |
Hb_005066_040 |
0.0851301765 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 4 |
Hb_000260_520 |
0.0877988602 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_001186_040 |
0.0954209539 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 6 |
Hb_005000_150 |
0.1000170576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008695_060 |
0.101133629 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_008705_080 |
0.1025453857 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 9 |
Hb_140595_020 |
0.1060664383 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |
| 10 |
Hb_009315_010 |
0.1069444991 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 11 |
Hb_001366_360 |
0.1090454052 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_001163_010 |
0.1093597259 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 13 |
Hb_000029_210 |
0.1095285545 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_001123_300 |
0.1100307911 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 15 |
Hb_005062_100 |
0.1110099154 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000427_020 |
0.1121324355 |
- |
- |
PsbA, partial [Dianthus pygmaeus] |
| 17 |
Hb_002170_030 |
0.1130603731 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 18 |
Hb_026198_080 |
0.1136946911 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 19 |
Hb_000462_040 |
0.1137769639 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 20 |
Hb_029587_010 |
0.1138657471 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |