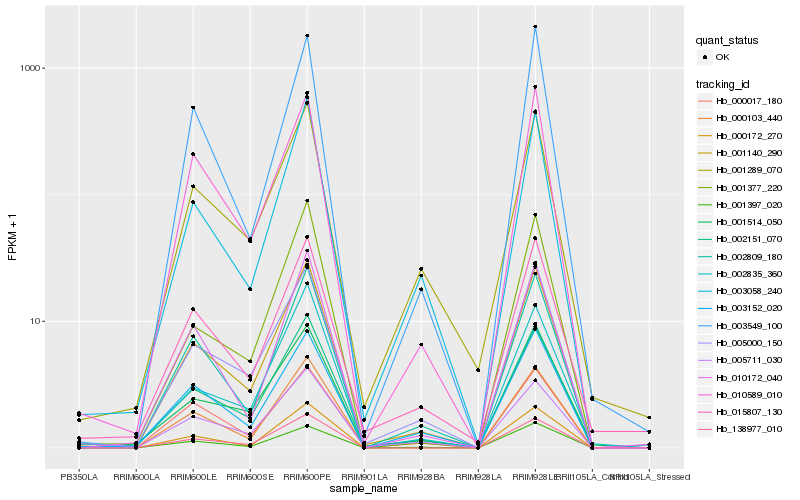
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001140_290 |
0.0 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 2 |
Hb_002151_070 |
0.040581112 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 2-like [Jatropha curcas] |
| 3 |
Hb_005000_150 |
0.0546858171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_138977_010 |
0.0649956823 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_002809_180 |
0.0651904834 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 6 |
Hb_000103_440 |
0.0675853203 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 7 |
Hb_010589_010 |
0.0745352235 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 8 |
Hb_001397_020 |
0.0755908209 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 9 |
Hb_000172_270 |
0.0791194126 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 10 |
Hb_005711_030 |
0.0802534849 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 11 |
Hb_003058_240 |
0.0805882824 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 12 |
Hb_003152_020 |
0.0822032967 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 13 |
Hb_015807_130 |
0.0849284808 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001289_070 |
0.0855698256 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_010172_040 |
0.0872541801 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001514_050 |
0.0873575468 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 17 |
Hb_003549_100 |
0.092387997 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 18 |
Hb_000017_180 |
0.0933982537 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 19 |
Hb_002835_360 |
0.0938041589 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 20 |
Hb_001377_220 |
0.0948228599 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |