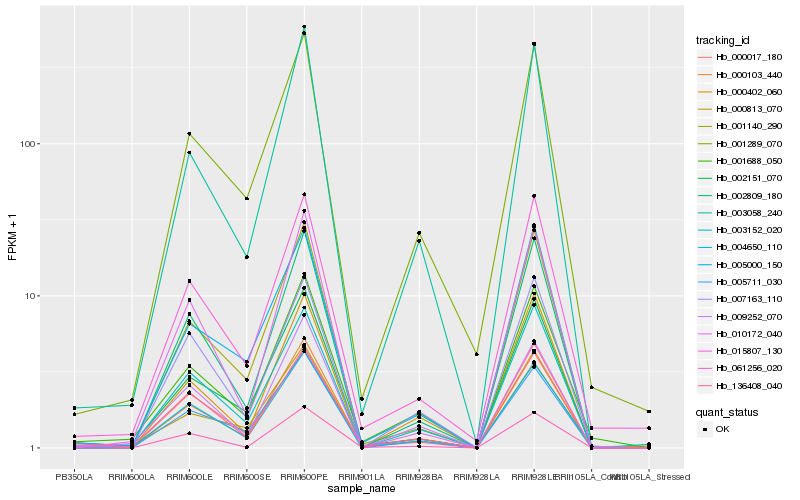
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_440 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 2 |
Hb_003058_240 |
0.0650410354 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 3 |
Hb_001140_290 |
0.0675853203 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 4 |
Hb_002151_070 |
0.0676035217 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 2-like [Jatropha curcas] |
| 5 |
Hb_002809_180 |
0.0727132072 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 6 |
Hb_005711_030 |
0.0743959955 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 7 |
Hb_003152_020 |
0.0798179255 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 8 |
Hb_001289_070 |
0.0811203883 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_001688_050 |
0.0814783783 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 10 |
Hb_061256_020 |
0.0847876987 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_136408_040 |
0.0861714652 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 12 |
Hb_004650_110 |
0.0869375684 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 13 |
Hb_015807_130 |
0.0878918401 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000813_070 |
0.0904687402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005000_150 |
0.0911583445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000017_180 |
0.0916288994 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 17 |
Hb_007163_110 |
0.0925132069 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 18 |
Hb_000402_060 |
0.0946183204 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 19 |
Hb_010172_040 |
0.0955959684 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_009252_070 |
0.0968654954 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |