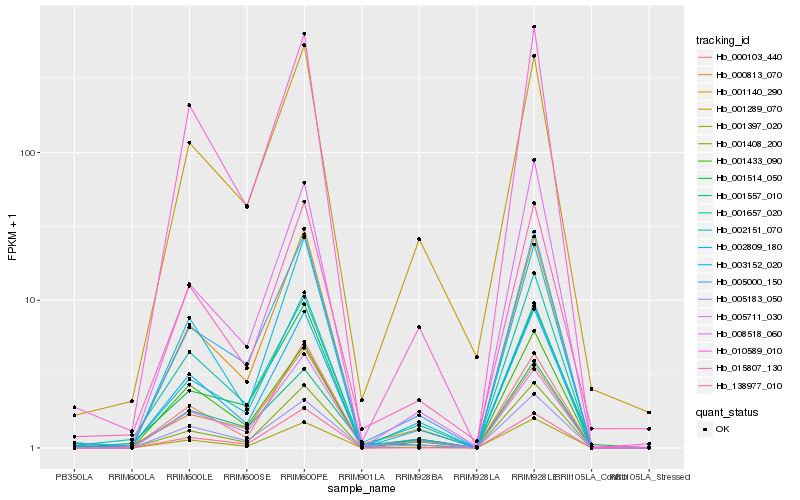
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005000_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001140_290 |
0.0546858171 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 3 |
Hb_002151_070 |
0.0590259412 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 2-like [Jatropha curcas] |
| 4 |
Hb_001514_050 |
0.0688986915 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 5 |
Hb_003152_020 |
0.0721333861 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 6 |
Hb_001397_020 |
0.0738595948 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 7 |
Hb_001289_070 |
0.0783514122 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 8 |
Hb_010589_010 |
0.0813920184 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 9 |
Hb_005183_050 |
0.0823245061 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 10 |
Hb_015807_130 |
0.0876379235 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001408_200 |
0.0889354284 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 12 |
Hb_005711_030 |
0.0889475028 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 13 |
Hb_000103_440 |
0.0911583445 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 14 |
Hb_001433_090 |
0.0930957891 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 15 |
Hb_008518_060 |
0.095151172 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 16 |
Hb_001657_020 |
0.0954484338 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 17 |
Hb_002809_180 |
0.0959782743 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 18 |
Hb_001557_010 |
0.095982282 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 19 |
Hb_000813_070 |
0.0962107236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_138977_010 |
0.0963342327 |
- |
- |
unnamed protein product [Vitis vinifera] |