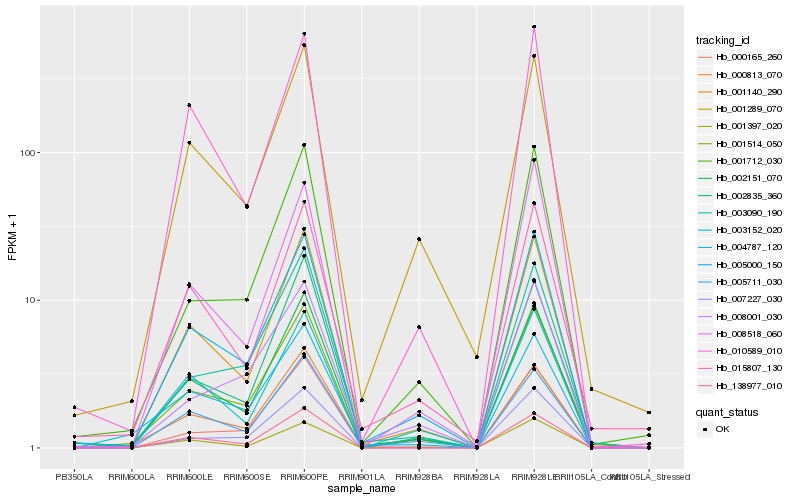
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_050 |
0.0 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 2 |
Hb_005000_150 |
0.0688986915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002151_070 |
0.0822003171 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 2-like [Jatropha curcas] |
| 4 |
Hb_001140_290 |
0.0873575468 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 5 |
Hb_001712_030 |
0.0913459756 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 6 |
Hb_003090_190 |
0.0919372111 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 7 |
Hb_001289_070 |
0.0919916626 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 8 |
Hb_007227_030 |
0.1004549986 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 9 |
Hb_002835_360 |
0.1011002454 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 10 |
Hb_138977_010 |
0.1011844158 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000165_260 |
0.1026303193 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 12 |
Hb_015807_130 |
0.1026997924 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000813_070 |
0.1033414832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003152_020 |
0.1037941728 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 15 |
Hb_004787_120 |
0.1050267051 |
- |
- |
PREDICTED: transcription factor TCP14 [Jatropha curcas] |
| 16 |
Hb_001397_020 |
0.1052571553 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 17 |
Hb_008001_030 |
0.1059520318 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_005711_030 |
0.1110995233 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 19 |
Hb_010589_010 |
0.1120035392 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 20 |
Hb_008518_060 |
0.1133765659 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |