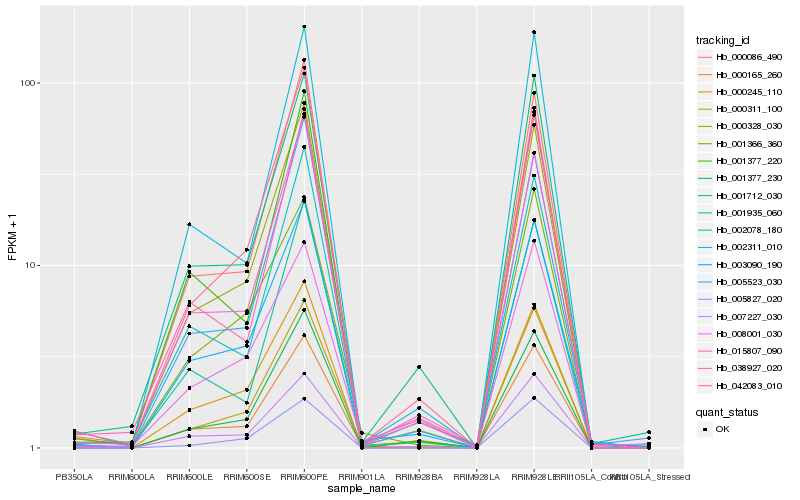
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 2 |
Hb_003090_190 |
0.0465085428 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 3 |
Hb_000328_030 |
0.0519676366 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 4 |
Hb_000311_100 |
0.0591437768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007227_030 |
0.0598489732 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 6 |
Hb_001712_030 |
0.0702800509 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 7 |
Hb_000086_490 |
0.0737956835 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 8 |
Hb_002311_010 |
0.0758186899 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 9 |
Hb_001377_220 |
0.0799266187 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 10 |
Hb_002078_180 |
0.0802229375 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 11 |
Hb_001377_230 |
0.0821648809 |
- |
- |
PREDICTED: uncharacterized protein LOC105135159 [Populus euphratica] |
| 12 |
Hb_001935_060 |
0.0825928508 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 13 |
Hb_015807_090 |
0.0840089159 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 14 |
Hb_005523_030 |
0.0873490048 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 15 |
Hb_038927_020 |
0.0894914253 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 16 |
Hb_001366_360 |
0.0896705154 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_008001_030 |
0.0906186175 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_042083_010 |
0.0911100259 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Jatropha curcas] |
| 19 |
Hb_005827_020 |
0.0924982117 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 20 |
Hb_000245_110 |
0.0929361795 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |