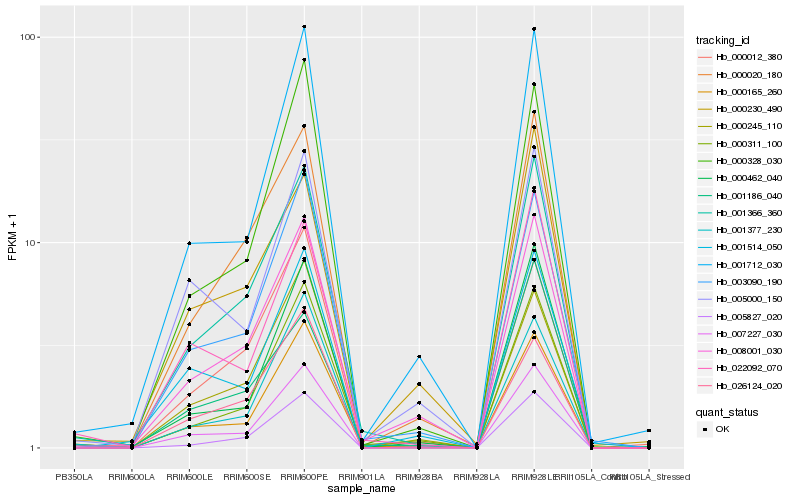
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_360 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_000020_180 |
0.0678661085 |
- |
- |
laccase, putative [Ricinus communis] |
| 3 |
Hb_008001_030 |
0.0808392019 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_007227_030 |
0.0834016187 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 5 |
Hb_000165_260 |
0.0896705154 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 6 |
Hb_000311_100 |
0.0972658417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005827_020 |
0.097968878 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 8 |
Hb_003090_190 |
0.1002205473 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 9 |
Hb_000328_030 |
0.1078586517 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 10 |
Hb_001712_030 |
0.1079796194 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 11 |
Hb_022092_070 |
0.1090454052 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.5 [Jatropha curcas] |
| 12 |
Hb_000462_040 |
0.1115684834 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 13 |
Hb_000012_380 |
0.1184989258 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000245_110 |
0.1199620719 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 15 |
Hb_001377_230 |
0.1202113676 |
- |
- |
PREDICTED: uncharacterized protein LOC105135159 [Populus euphratica] |
| 16 |
Hb_001186_040 |
0.1204034289 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 17 |
Hb_005000_150 |
0.1266026276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001514_050 |
0.1271448292 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 19 |
Hb_026124_020 |
0.1289347511 |
- |
- |
PREDICTED: uncharacterized protein LOC105644241 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000230_490 |
0.1304189963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |